Protein–protein interaction on:
[Wikipedia]
[Google]
[Amazon]
 Protein–protein interactions (PPIs) are physical contacts of high specificity established between two or more
Protein–protein interactions (PPIs) are physical contacts of high specificity established between two or more

 The molecular structures of many protein complexes have been unlocked by the technique of
The molecular structures of many protein complexes have been unlocked by the technique of


 Many computational methods have been suggested and reviewed for predicting protein-protein interactions. Prediction approaches can be grouped into categories based on predictive evidence: protein sequence,
Many computational methods have been suggested and reviewed for predicting protein-protein interactions. Prediction approaches can be grouped into categories based on predictive evidence: protein sequence,
 Information found in PPIs databases supports the construction of interaction networks. Although the PPI network of a given query protein can be represented in textbooks, diagrams of whole cell PPIs are frankly complex and difficult to generate.
One example of a manually produced molecular interaction map is the Kurt Kohn's 1999 map of cell cycle control. Drawing on Kohn's map, Schwikowski et al. in 2000 published a paper on PPIs in yeast, linking 1,548 interacting proteins determined by two-hybrid screening. They used a layered graph drawing method to find an initial placement of the nodes and then improved the layout using a force-based algorithm.
Bioinformatic tools have been developed to simplify the difficult task of visualizing molecular interaction networks and complement them with other types of data. For instance,
Information found in PPIs databases supports the construction of interaction networks. Although the PPI network of a given query protein can be represented in textbooks, diagrams of whole cell PPIs are frankly complex and difficult to generate.
One example of a manually produced molecular interaction map is the Kurt Kohn's 1999 map of cell cycle control. Drawing on Kohn's map, Schwikowski et al. in 2000 published a paper on PPIs in yeast, linking 1,548 interacting proteins determined by two-hybrid screening. They used a layered graph drawing method to find an initial placement of the nodes and then improved the layout using a force-based algorithm.
Bioinformatic tools have been developed to simplify the difficult task of visualizing molecular interaction networks and complement them with other types of data. For instance,
 Protein–protein interactions often result in one of the interacting proteins either being 'activated' or 'repressed'. Such effects can be indicated in a PPI network by "signs" (e.g. "activation" or "inhibition"). Although such attributes have been added to networks for a long time, Vinayagam et al. (2014) coined the term ''Signed network'' for them. Signed networks are often expressed by labeling the interaction as either positive or negative. A positive interaction is one where the interaction results in one of the proteins being activated. Conversely, a negative interaction indicates that one of the proteins being inactivated.
Protein–protein interaction networks are often constructed as a result of lab experiments such as yeast two-hybrid screens or 'affinity purification and subsequent mass spectrometry techniques. However these methods do not provide the layer of information needed in order to determine what type of interaction is present in order to be able to attribute signs to the network diagrams.
Protein–protein interactions often result in one of the interacting proteins either being 'activated' or 'repressed'. Such effects can be indicated in a PPI network by "signs" (e.g. "activation" or "inhibition"). Although such attributes have been added to networks for a long time, Vinayagam et al. (2014) coined the term ''Signed network'' for them. Signed networks are often expressed by labeling the interaction as either positive or negative. A positive interaction is one where the interaction results in one of the proteins being activated. Conversely, a negative interaction indicates that one of the proteins being inactivated.
Protein–protein interaction networks are often constructed as a result of lab experiments such as yeast two-hybrid screens or 'affinity purification and subsequent mass spectrometry techniques. However these methods do not provide the layer of information needed in order to determine what type of interaction is present in order to be able to attribute signs to the network diagrams.
Protein-Protein Interaction Databases
Library of Modulators of Protein–Protein Interactions (PPI)
* * {{DEFAULTSORT:Protein-Protein Interaction Proteomics Signal transduction Biophysics Biochemistry methods Biotechnology Quantum biochemistry Protein-protein interaction assays Protein complexes
 Protein–protein interactions (PPIs) are physical contacts of high specificity established between two or more
Protein–protein interactions (PPIs) are physical contacts of high specificity established between two or more protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, res ...
molecules as a result of biochemical events steered by interactions that include electrostatic forces
Coulomb's inverse-square law, or simply Coulomb's law, is an experimental law of physics that quantifies the amount of force between two stationary, electrically charged particles. The electric force between charged bodies at rest is convention ...
, hydrogen bond
In chemistry, a hydrogen bond (or H-bond) is a primarily electrostatic force of attraction between a hydrogen (H) atom which is covalently bound to a more electronegative "donor" atom or group (Dn), and another electronegative atom bearing a l ...
ing and the hydrophobic effect
The hydrophobic effect is the observed tendency of nonpolar substances to aggregate in an aqueous solution and exclude water molecules. The word hydrophobic literally means "water-fearing", and it describes the segregation of water and nonpolar ...
. Many are physical contacts with molecular associations between chains that occur in a cell or in a living organism in a specific biomolecular context.
Proteins rarely act alone as their functions tend to be regulated. Many molecular processes within a cell are carried out by molecular machine
A molecular machine, nanite, or nanomachine is a molecular component that produces quasi-mechanical movements (output) in response to specific stimuli (input). In cellular biology, macromolecular machines frequently perform tasks essential for l ...
s that are built from numerous protein components organized by their PPIs. These physiological interactions make up the so-called interactomics of the organism, while aberrant PPIs are the basis of multiple aggregation-related diseases, such as Creutzfeldt–Jakob and Alzheimer's disease
Alzheimer's disease (AD) is a neurodegenerative disease that usually starts slowly and progressively worsens. It is the cause of 60–70% of cases of dementia. The most common early symptom is difficulty in remembering recent events. As ...
s.
PPIs have been studied with many methods and from different perspectives: biochemistry
Biochemistry or biological chemistry is the study of chemical processes within and relating to living organisms. A sub-discipline of both chemistry and biology, biochemistry may be divided into three fields: structural biology, enzymology and ...
, quantum chemistry
Quantum chemistry, also called molecular quantum mechanics, is a branch of physical chemistry focused on the application of quantum mechanics to chemical systems, particularly towards the quantum-mechanical calculation of electronic contributions ...
, molecular dynamics
Molecular dynamics (MD) is a computer simulation method for analyzing the physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a fixed period of time, giving a view of the dynamic "evolution" of th ...
, signal transduction
Signal transduction is the process by which a chemical or physical signal is transmitted through a cell as a series of molecular events, most commonly protein phosphorylation catalyzed by protein kinases, which ultimately results in a cellula ...
, among others. All this information enables the creation of large protein interaction networks – similar to metabolic
Metabolism (, from el, μεταβολή ''metabolē'', "change") is the set of life-sustaining chemical reactions in organisms. The three main functions of metabolism are: the conversion of the energy in food to energy available to run cell ...
or genetic/epigenetic networks – that empower the current knowledge on biochemical cascades and molecular etiology of disease, as well as the discovery of putative protein targets of therapeutic interest.
Examples
Electron transfer protein
The electron ( or ) is a subatomic particle with a negative one elementary electric charge. Electrons belong to the first generation of the lepton particle family,
and are generally thought to be elementary particles because they have no ...
s
In many metabolic reactions, a protein that acts as an electron carrier binds to an enzyme that acts its reductase
A reductase is an enzyme that catalyzes a reduction reaction.
Examples
* 5α-Reductase
* 5β-Reductase
* Dihydrofolate reductase
* HMG-CoA reductase
* Methemoglobin reductase
* Ribonucleotide reductase
* Thioredoxin reductase
* ''E. coli' ...
. After it receives an electron, it dissociates and then binds to the next enzyme that acts its oxidase (i.e. an acceptor of the electron). These interactions between proteins are dependent on highly specific binding between proteins to ensure efficient electron transfer. Examples: mitochondrial oxidative phosphorylation chain system components cytochrome c-reductase / cytochrome c
The cytochrome complex, or cyt ''c'', is a small hemeprotein found loosely associated with the inner membrane of the mitochondrion. It belongs to the cytochrome c family of proteins and plays a major role in cell apoptosis. Cytochrome c is hig ...
/ cytochrome c oxidase; microsomal and mitochondrial P450 systems.
In the case of the mitochondrial P450 systems, the specific residues involved in the binding of the electron transfer protein adrenodoxin to its reductase were identified as two basic Arg residues on the surface of the reductase and two acidic Asp residues on the adrenodoxin.
More recent work on the phylogeny of the reductase has shown that these residues involved in protein-protein interactions have been conserved throughout the evolution of this enzyme.
Signal transduction
Signal transduction is the process by which a chemical or physical signal is transmitted through a cell as a series of molecular events, most commonly protein phosphorylation catalyzed by protein kinases, which ultimately results in a cellula ...
The activity of the cell is regulated by extracellular signals. Signal propagation inside and/or along the interior of cells depends on PPIs between the various signaling molecules. The recruitment of signaling pathways through PPIs is called signal transduction
Signal transduction is the process by which a chemical or physical signal is transmitted through a cell as a series of molecular events, most commonly protein phosphorylation catalyzed by protein kinases, which ultimately results in a cellula ...
and plays a fundamental role in many biological processes and in many diseases including Parkinson's disease
Parkinson's disease (PD), or simply Parkinson's, is a long-term degenerative disorder of the central nervous system that mainly affects the motor system. The symptoms usually emerge slowly, and as the disease worsens, non-motor symptoms beco ...
and cancer.
Membrane transport
In cellular biology, membrane transport refers to the collection of mechanisms that regulate the passage of solutes such as ions and small molecules through biological membranes, which are lipid bilayers that contain proteins embedded in them. The ...
A protein may be carrying another protein (for example, from cytoplasm
In cell biology, the cytoplasm is all of the material within a eukaryotic cell, enclosed by the cell membrane, except for the cell nucleus. The material inside the nucleus and contained within the nuclear membrane is termed the nucleoplasm. ...
to nucleus
Nucleus ( : nuclei) is a Latin word for the seed inside a fruit. It most often refers to:
* Atomic nucleus, the very dense central region of an atom
*Cell nucleus, a central organelle of a eukaryotic cell, containing most of the cell's DNA
Nucl ...
or vice versa in the case of the nuclear pore
A nuclear pore is a part of a large complex of proteins, known as a nuclear pore complex that spans the nuclear envelope, which is the double membrane surrounding the eukaryotic cell nucleus. There are approximately 1,000 nuclear pore comple ...
importins).
Cell metabolism
Metabolism (, from el, μεταβολή ''metabolē'', "change") is the set of life-sustaining chemical reactions in organisms. The three main functions of metabolism are: the conversion of the energy in food to energy available to run c ...
In many biosynthetic processes enzymes
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. ...
interact with each other to produce small compounds or other macromolecules.
Muscle contraction
Muscle contraction is the activation of tension-generating sites within muscle cells. In physiology, muscle contraction does not necessarily mean muscle shortening because muscle tension can be produced without changes in muscle length, such a ...
Physiology of muscle contraction
Muscle contraction is the activation of tension-generating sites within muscle cells. In physiology, muscle contraction does not necessarily mean muscle shortening because muscle tension can be produced without changes in muscle length, such a ...
involves several interactions. Myosin
Myosins () are a superfamily of motor proteins best known for their roles in muscle contraction and in a wide range of other motility processes in eukaryotes. They are ATP-dependent and responsible for actin-based motility.
The first myosin (M ...
filaments act as molecular motor
Molecular motors are natural (biological) or artificial molecular machines that are the essential agents of movement in living organisms. In general terms, a motor is a device that consumes energy in one form and converts it into motion or mec ...
s and by binding to actin
Actin is a family of globular multi-functional proteins that form microfilaments in the cytoskeleton, and the thin filaments in muscle fibrils. It is found in essentially all eukaryotic cells, where it may be present at a concentration of ov ...
enables filament sliding. Furthermore, members of the skeletal muscle
Skeletal muscles (commonly referred to as muscles) are organs of the vertebrate muscular system and typically are attached by tendons to bones of a skeleton. The muscle cells of skeletal muscles are much longer than in the other types of m ...
lipid droplet-associated proteins family associate with other proteins, as activator of adipose triglyceride lipase
Adipose triglyceride lipase, also known as patatin-like phospholipase domain-containing protein 2 and ATGL, is an enzyme that in humans is encoded by the ''PNPLA2'' gene. ATGL catalyses the first reaction of lipolysis, where triacylglycerols a ...
and its coactivator comparative gene identification-58, to regulate lipolysis
Lipolysis is the metabolic pathway through which lipid triglycerides are hydrolyzed into a glycerol and free fatty acids. It is used to mobilize stored energy during fasting or exercise, and usually occurs in fat adipocytes. The most important ...
in skeletal muscle
Types
To describe the types of protein–protein interactions (PPIs) it is important to consider that proteins can interact in a "transient" way (to produce some specific effect in a short time, like a signal transduction) or to interact with other proteins in a "stable" way to form complexes that become molecular machines within the living systems. A protein complex assembly can result in the formation of homo-oligomeric or hetero-oligomeric complexes. In addition to the conventional complexes, as enzyme-inhibitor and antibody-antigen, interactions can also be established between domain-domain and domain-peptide. Another important distinction to identify protein-protein interactions is the way they have been determined, since there are techniques that measure direct physical interactions between protein pairs, named “binary” methods, while there are other techniques that measure physical interactions among groups of proteins, without pairwise determination of protein partners, named “co-complex” methods.Homo-oligomers vs. hetero-oligomers
Homo-oligomers are macromolecular complexes constituted by only one type ofprotein subunit
In structural biology, a protein subunit is a polypeptide chain or single protein molecule that assembles (or "''coassembles''") with others to form a protein complex.
Large assemblies of proteins such as viruses often use a small number of t ...
. Protein subunits assembly is guided by the establishment of non-covalent interactions in the quaternary structure of the protein. Disruption of homo-oligomers in order to return to the initial individual monomers
In chemistry, a monomer ( ; ''mono-'', "one" + '' -mer'', "part") is a molecule that can react together with other monomer molecules to form a larger polymer chain or three-dimensional network in a process called polymerization.
Classification
Mo ...
often requires denaturation of the complex. Several enzymes
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. ...
, carrier proteins
A membrane transport protein (or simply transporter) is a membrane protein involved in the movement of ions, small molecules, and macromolecules, such as another protein, across a biological membrane. Transport proteins are integral transmembrane ...
, scaffolding proteins, and transcriptional regulatory factors carry out their functions as homo-oligomers.
Distinct protein subunits interact in hetero-oligomers, which are essential to control several cellular functions. The importance of the communication between heterologous proteins is even more evident during cell signaling events and such interactions are only possible due to structural domains within the proteins (as described below).
Stable interactions vs. transient interactions
Stable interactions involve proteins that interact for a long time, taking part of permanent complexes as subunits, in order to carry out functional roles. These are usually the case of homo-oligomers (e.g.cytochrome c
The cytochrome complex, or cyt ''c'', is a small hemeprotein found loosely associated with the inner membrane of the mitochondrion. It belongs to the cytochrome c family of proteins and plays a major role in cell apoptosis. Cytochrome c is hig ...
), and some hetero-oligomeric proteins, as the subunits of ATPase
ATPases (, Adenosine 5'-TriPhosphatase, adenylpyrophosphatase, ATP monophosphatase, triphosphatase, SV40 T-antigen, ATP hydrolase, complex V (mitochondrial electron transport), (Ca2+ + Mg2+)-ATPase, HCO3−-ATPase, adenosine triphosphatase) are ...
. On the other hand, a protein may interact briefly and in a reversible manner with other proteins in only certain cellular contexts – cell type
A cell type is a classification used to identify cells that share morphological or phenotypical features. A multicellular organism may contain cells of a number of widely differing and specialized cell types, such as muscle cells and skin cell ...
, cell cycle stage, external factors, presence of other binding proteins, etc. – as it happens with most of the proteins involved in biochemical cascades. These are called transient interactions. For example, some G protein-coupled receptors only transiently bind to Gi/o proteins when they are activated by extracellular ligands, while some Gq-coupled receptors, such as muscarinic receptor M3, pre-couple with Gq proteins prior to the receptor-ligand binding. Interactions between intrinsically disordered protein regions to globular protein domains (i.e. MoRFs Molecular recognition features (MoRFs) are small (10-70 residues) intrinsically disordered regions in proteins that undergo a disorder-to-order transition upon binding to their partners. MoRFs are implicated in protein-protein interactions, which ...
) are transient interactions.
Covalent vs. non-covalent
Covalent interactions are those with the strongest association and are formed by disulphide bonds or electron sharing. While rare, these interactions are determinant in some posttranslational modifications, asubiquitination
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Fo ...
and SUMOylation
In molecular biology, SUMO (Small Ubiquitin-like Modifier) proteins are a family of small proteins that are covalently attached to and detached from other proteins in cells to modify their function. This process is called SUMOylation (sometimes w ...
. Non-covalent bonds are usually established during transient interactions by the combination of weaker bonds, such as hydrogen bonds
In chemistry, a hydrogen bond (or H-bond) is a primarily electrostatic force of attraction between a hydrogen (H) atom which is covalently bound to a more electronegative "donor" atom or group (Dn), and another electronegative atom bearing a l ...
, ionic interactions, Van der Waals force
In molecular physics, the van der Waals force is a distance-dependent interaction between atoms or molecules. Unlike ionic or covalent bonds, these attractions do not result from a chemical electronic bond; they are comparatively weak and ...
s, or hydrophobic bonds.
Role of water
Water molecules play a significant role in the interactions between proteins. The crystal structures of complexes, obtained at high resolution from different but homologous proteins, have shown that some interface water molecules are conserved between homologous complexes. The majority of the interface water molecules make hydrogen bonds with both partners of each complex. Some interface amino acid residues or atomic groups of one protein partner engage in both direct and water mediated interactions with the other protein partner. Doubly indirect interactions, mediated by two water molecules, are more numerous in the homologous complexes of low affinity. Carefully conducted mutagenesis experiments, e.g. changing a tyrosine residue into a phenylalanine, have shown that water mediated interactions can contribute to the energy of interaction. Thus, water molecules may facilitate the interactions and cross-recognitions between proteins.Structure

 The molecular structures of many protein complexes have been unlocked by the technique of
The molecular structures of many protein complexes have been unlocked by the technique of X-ray crystallography
X-ray crystallography is the experimental science determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam of incident X-rays to diffract into many specific directions. By measuring the angles ...
. The first structure to be solved by this method was that of sperm whale
The sperm whale or cachalot (''Physeter macrocephalus'') is the largest of the toothed whales and the largest toothed predator. It is the only living member of the genus ''Physeter'' and one of three extant species in the sperm whale famil ...
myoglobin
Myoglobin (symbol Mb or MB) is an iron- and oxygen-binding protein found in the cardiac and skeletal muscle tissue of vertebrates in general and in almost all mammals. Myoglobin is distantly related to hemoglobin. Compared to hemoglobin, myoglob ...
by Sir John Cowdery Kendrew. In this technique the angles and intensities of a beam of X-rays diffracted by crystalline atoms are detected in a film, thus producing a three-dimensional picture of the density of electrons within the crystal.
Later, nuclear magnetic resonance
Nuclear magnetic resonance (NMR) is a physical phenomenon in which nuclei in a strong constant magnetic field are perturbed by a weak oscillating magnetic field (in the near field) and respond by producing an electromagnetic signal with a ...
also started to be applied with the aim of unravelling the molecular structure of protein complexes. One of the first examples was the structure of calmodulin-binding domains bound to calmodulin
Calmodulin (CaM) (an abbreviation for calcium-modulated protein) is a multifunctional intermediate calcium-binding messenger protein expressed in all eukaryotic cells. It is an intracellular target of the secondary messenger Ca2+, and the bin ...
. This technique is based on the study of magnetic properties of atomic nuclei, thus determining physical and chemical properties of the correspondent atoms or the molecules. Nuclear magnetic resonance is advantageous for characterizing weak PPIs.
Domains
Proteins hold structural domains that allow their interaction with and bind to specific sequences on other proteins: * ''Src homology 2 (SH2) domain '' :SH2 domains are structurally composed by three-stranded twisted beta sheet sandwiched flanked by two alpha-helices. The existence of a deep binding pocket with high affinity forphosphotyrosine
-Tyrosine or tyrosine (symbol Tyr or Y) or 4-hydroxyphenylalanine is one of the 20 standard amino acids that are used by cells to synthesize proteins. It is a non-essential amino acid with a polar side group. The word "tyrosine" is from the G ...
, but not for phosphoserine or phosphothreonine, is essential for the recognition of tyrosine phosphorylated proteins, mainly autophosphorylated growth factor receptors. Growth factor receptor binding proteins and phospholipase C
Phospholipase C (PLC) is a class of membrane-associated enzymes that cleave phospholipids just before the phosphate group (see figure). It is most commonly taken to be synonymous with the human forms of this enzyme, which play an important role ...
γ are examples of proteins that have SH2 domains.
* ''Src homology 3 (SH3) domain ''
:Structurally, SH3 domains are constituted by a beta barrel formed by two orthogonal beta sheets and three anti-parallel beta strands. These domains recognize proline
Proline (symbol Pro or P) is an organic acid classed as a proteinogenic amino acid (used in the biosynthesis of proteins), although it does not contain the amino group but is rather a secondary amine. The secondary amine nitrogen is in the p ...
enriched sequences, as polyproline type II helical structure (PXXP motifs) in cell signaling proteins like protein tyrosine kinases and the growth factor receptor bound protein 2 ( Grb2).
* ''Phosphotyrosine-binding (PTB) domain ''
:PTB domains interact with sequences that contain a phosphotyrosine group. These domains can be found in the insulin receptor substrate Insulin receptor substrate (IRS) is an important ligand in the insulin response of human cells.
IRS-1
Insulin receptor substrate 1 (IRS-1) is a signaling adapter protein that in humans is encoded by the ''IRS-1'' gene. It is a 131 kDa protein w ...
.
* ''LIM domain ''
:LIM domains were initially identified in three homeodomain transcription factors (lin11, is11, and mec3). In addition to this homeodomain proteins and other proteins involved in development, LIM domains have also been identified in non-homeodomain proteins with relevant roles in cellular differentiation
Cellular differentiation is the process in which a stem cell alters from one type to a differentiated one. Usually, the cell changes to a more specialized type. Differentiation happens multiple times during the development of a multicellular ...
, association with cytoskeleton
The cytoskeleton is a complex, dynamic network of interlinking protein filaments present in the cytoplasm of all cells, including those of bacteria and archaea. In eukaryotes, it extends from the cell nucleus to the cell membrane and is co ...
and senescence
Senescence () or biological aging is the gradual deterioration of functional characteristics in living organisms. The word ''senescence'' can refer to either cellular senescence or to senescence of the whole organism. Organismal senescence invol ...
. These domains contain a tandem cysteine-rich Zn2+-finger motif and embrace the consensus sequence
In molecular biology and bioinformatics, the consensus sequence (or canonical sequence) is the calculated order of most frequent residues, either nucleotide or amino acid, found at each position in a sequence alignment. It serves as a simplified r ...
CX2CX16-23HX2CX2CX2CX16-21CX2C/H/D. LIM domains bind to PDZ domains, bHLH transcription factors, and other LIM domains.
* ''Sterile alpha motif (SAM) domain ''
:SAM domains are composed by five helices forming a compact package with a conserved hydrophobic core. These domains, which can be found in the Eph receptor
Eph receptors (Ephs, after erythropoietin-producing human hepatocellular receptors) are a group of receptors that are activated in response to binding with Eph receptor-interacting proteins (Ephrins). Ephs form the largest known subfamily of re ...
and the stromal interaction molecule ( STIM) for example, bind to non-SAM domain-containing proteins and they also appear to have the ability to bind RNA.
* ''PDZ domain ''
:PDZ domains were first identified in three guanylate kinases: PSD-95, DlgA and ZO-1. These domains recognize carboxy-terminal tri-peptide motifs (S/TXV), other PDZ domains or LIM domains and bind them through a short peptide sequence that has a C-terminal hydrophobic residue. Some of the proteins identified as having PDZ domains are scaffolding proteins or seem to be involved in ion receptor assembling and receptor-enzyme complexes formation.
* ''FERM domain ''
:FERM domains contain basic residues capable of binding PtdIns(4,5)P2. Talin Talin may refer to:
Places
*Talin, Armenia, a city
* Tálín, a municipality and village in the Czech Republic
*Tallinn, capital of Estonia
* Talin, Iran, a village in West Azerbaijan Province
*Talin, Syria, a village in Tartus Governorate
Other
* ...
and focal adhesion kinase (FAK) are two of the proteins that present FERM domains.
* ''Calponin homology (CH) domain ''
:CH domains are mainly present in cytoskeletal proteins as parvin.
* ''Pleckstrin homology domain ''
:Pleckstrin homology domains bind to phosphoinositides and acid domains in signaling proteins.
* ''WW domain ''
:WW domains bind to proline enriched sequences.
* ''WSxWS motif ''
:Found in cytokine receptors
Properties of the interface
The study of the molecular structure can give fine details about the interface that enables the interaction between proteins. When characterizing PPI interfaces it is important to take into account the type of complex. Parameters evaluated include size (measured in absolute dimensions Å2 or in solvent-accessible surface area (SASA)), shape, complementarity between surfaces, residue interface propensities, hydrophobicity, segmentation and secondary structure, and conformational changes on complex formation. The great majority of PPI interfaces reflects the composition of protein surfaces, rather than the protein cores, in spite of being frequently enriched in hydrophobic residues, particularly in aromatic residues. PPI interfaces are dynamic and frequently planar, although they can be globular and protruding as well. Based on three structures –insulin
Insulin (, from Latin ''insula'', 'island') is a peptide hormone produced by beta cells of the pancreatic islets encoded in humans by the ''INS'' gene. It is considered to be the main anabolic hormone of the body. It regulates the metabolism ...
dimer, trypsin
Trypsin is an enzyme in the first section of the small intestine that starts the digestion of protein molecules by cutting these long chains of amino acids into smaller pieces. It is a serine protease from the PA clan superfamily, found in the d ...
-pancreatic trypsin inhibitor complex, and oxyhaemoglobin – Cyrus Chothia and Joel Janin found that between 1,130 and 1,720 Å2 of surface area was removed from contact with water indicating that hydrophobicity is a major factor of stabilization of PPIs. Later studies refined the buried surface area of the majority of interactions to 1,600±350 Å2. However, much larger interaction interfaces were also observed and were associated with significant changes in conformation of one of the interaction partners. PPIs interfaces exhibit both shape and electrostatic complementarity.
Regulation
* Protein concentration, which in turn are affected by expression levels and degradation rates; * Protein affinity for proteins or other binding ligands; * Ligands concentrations ( substrates,ions
An ion () is an atom or molecule with a net electrical charge.
The charge of an electron is considered to be negative by convention and this charge is equal and opposite to the charge of a proton, which is considered to be positive by conven ...
, etc.);
* Presence of other proteins
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, respo ...
, nucleic acids
Nucleic acids are biopolymers, macromolecules, essential to all known forms of life. They are composed of nucleotides, which are the monomers made of three components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main cl ...
, and ions
An ion () is an atom or molecule with a net electrical charge.
The charge of an electron is considered to be negative by convention and this charge is equal and opposite to the charge of a proton, which is considered to be positive by conven ...
;
* Electric field
An electric field (sometimes E-field) is the physical field that surrounds electrically charged particles and exerts force on all other charged particles in the field, either attracting or repelling them. It also refers to the physical field ...
s around proteins.
* Occurrence of covalent modifications;
Experimental methods
There are a multitude of methods to detect them. Each of the approaches has its own strengths and weaknesses, especially with regard to thesensitivity and specificity
''Sensitivity'' and ''specificity'' mathematically describe the accuracy of a test which reports the presence or absence of a condition. Individuals for which the condition is satisfied are considered "positive" and those for which it is not are ...
of the method. The most conventional and widely used high-throughput methods are yeast two-hybrid screening and affinity purification
Affinity chromatography is a method of separating a biomolecule from a mixture, based on a highly specific macromolecular binding interaction between the biomolecule and another substance. The specific type of binding interaction depends on the ...
coupled to mass spectrometry
Mass spectrometry (MS) is an analytical technique that is used to measure the mass-to-charge ratio of ions. The results are presented as a '' mass spectrum'', a plot of intensity as a function of the mass-to-charge ratio. Mass spectrometry is u ...
.
Yeast two-hybrid screening
This system was firstly described in 1989 by Fields and Song using ''Saccharomyces cerevisiae'' as biological model. Yeast two hybrid allows the identification of pairwise PPIs (binary method) ''in vivo'', in which the two proteins are tested for biophysically direct interaction. The Y2H is based on the functional reconstitution of the yeast transcription factor Gal4 and subsequent activation of a selective reporter such as His3. To test two proteins for interaction, two protein expression constructs are made: one protein (X) is fused to the Gal4 DNA-binding domain (DB) and a second protein (Y) is fused to the Gal4 activation domain (AD). In the assay, yeast cells are transformed with these constructs. Transcription of reporter genes does not occur unless bait (DB-X) and prey (AD-Y) interact with each other and form a functional Gal4 transcription factor. Thus, the interaction between proteins can be inferred by the presence of the products resultant of the reporter gene expression. In cases in which the reporter gene expresses enzymes that allow the yeast to synthesize essential amino acids or nucleotides, yeast growth under selective media conditions indicates that the two proteins tested are interacting. Recently, software to detect and prioritize protein interactions was published. Despite its usefulness, the yeast two-hybrid system has limitations. It uses yeast as main host system, which can be a problem when studying proteins that contain mammalian-specific post-translational modifications. The number of PPIs identified is usually low because of a high false negative rate; and, understatesmembrane proteins
Membrane proteins are common proteins that are part of, or interact with, biological membranes. Membrane proteins fall into several broad categories depending on their location. Integral membrane proteins are a permanent part of a cell membrane ...
, for example.
In initial studies that utilized Y2H, proper controls for false positives (e.g. when DB-X activates the reporter gene without the presence of AD-Y) were frequently not done, leading to a higher than normal false positive rate. An empirical framework must be implemented to control for these false positives. Limitations in lower coverage of membrane proteins have been overcoming by the emergence of yeast two-hybrid variants, such as the membrane yeast two-hybrid (MYTH) and the split-ubiquitin system, which are not limited to interactions that occur in the nucleus; and, the bacterial two-hybrid system, performed in bacteria;
Affinity purification coupled to mass spectrometry
Affinity purification coupled to mass spectrometry mostly detects stable interactions and thus better indicates functional in vivo PPIs. This method starts by purification of the tagged protein, which is expressed in the cell usually at ''in vivo'' concentrations, and its interacting proteins (affinity purification). One of the most advantageous and widely used methods to purify proteins with very low contaminating background is the tandem affinity purification, developed by Bertrand Seraphin and Matthias Mann and respective colleagues. PPIs can then be quantitatively and qualitatively analysed by mass spectrometry using different methods: chemical incorporation, biological or metabolic incorporation (SILAC), and label-free methods. Furthermore,network theory
Network theory is the study of graphs as a representation of either symmetric relations or asymmetric relations between discrete objects. In computer science and network science, network theory is a part of graph theory: a network can be de ...
has been used to study the whole set of identified protein-protein interactions in cells.
Nucleic acid programmable protein array (NAPPA)
This system was first developed by LaBaer and colleagues in 2004 by using in vitro transcription and translation system. They use DNA template encoding the gene of interest fused with GST protein, and it was immobilized in the solid surface. Anti-GST antibody and biotinylated plasmid DNA were bounded in aminopropyltriethoxysilane (APTES)-coated slide. BSA can improve the binding efficiency of DNA. Biotinylated plasmid DNA was bound by avidin. New protein was synthesized by using cell-free expression system i.e. rabbit reticulocyte lysate (RRL), and then the new protein was captured through anti-GST antibody bounded on the slide. To test protein-protein interaction, the targeted protein cDNA and query protein cDNA were immobilized in a same coated slide. By using in vitro transcription and translation system, targeted and query protein was synthesized by the same extract. The targeted protein was bound to array by antibody coated in the slide and query protein was used to probe the array. The query protein was tagged with hemagglutinin (HA) epitope. Thus, the interaction between the two proteins was visualized with the antibody against HA.Intragenic complementation
When multiple copies of a polypeptide encoded by agene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b ...
form a complex, this protein structure is referred to as a multimer. When a multimer is formed from polypeptides produced by two different mutant
In biology, and especially in genetics, a mutant is an organism or a new genetic character arising or resulting from an instance of mutation, which is generally an alteration of the DNA sequence of the genome or chromosome of an organism. It ...
allele
An allele (, ; ; modern formation from Greek ἄλλος ''állos'', "other") is a variation of the same sequence of nucleotides at the same place on a long DNA molecule, as described in leading textbooks on genetics and evolution.
::"The chrom ...
s of a particular gene, the mixed multimer may exhibit greater functional activity than the unmixed multimers formed by each of the mutants alone. In such a case, the phenomenon is referred to as intragenic complementation
Epistasis is a phenomenon in genetics in which the effect of a gene mutation is dependent on the presence or absence of mutations in one or more other genes, respectively termed modifier genes. In other words, the effect of the mutation is dep ...
(also called inter-allelic complementation). Intragenic complementation has been demonstrated in many different genes in a variety of organisms including the fungi '' Neurospora crassa'', ''Saccharomyces cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungus microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have b ...
'' and '' Schizosaccharomyces pombe''; the bacterium ''Salmonella
''Salmonella'' is a genus of rod-shaped (bacillus) Gram-negative bacteria of the family Enterobacteriaceae. The two species of ''Salmonella'' are '' Salmonella enterica'' and '' Salmonella bongori''. ''S. enterica'' is the type species and is ...
typhimurium''; the virus bacteriophage T4, an RNA virus and humans. In such studies, numerous mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, m ...
s defective in the same gene were often isolated and mapped in a linear order on the basis of recombination frequencies to form a genetic map
Genetic linkage is the tendency of DNA sequences that are close together on a chromosome to be inherited together during the meiosis phase of sexual reproduction. Two genetic markers that are physically near to each other are unlikely to be sep ...
of the gene. Separately, the mutants were tested in pairwise combinations to measure complementation. An analysis of the results from such studies led to the conclusion that intragenic complementation, in general, arises from the interaction of differently defective polypeptide monomers to form a multimer. Genes that encode multimer-forming polypeptides appear to be common. One interpretation of the data is that polypeptide monomers are often aligned in the multimer in such a way that mutant polypeptides defective at nearby sites in the genetic map tend to form a mixed multimer that functions poorly, whereas mutant polypeptides defective at distant sites tend to form a mixed multimer that functions more effectively. Direct interaction of two nascent proteins emerging from nearby ribosome
Ribosomes ( ) are macromolecular machines, found within all cells, that perform biological protein synthesis (mRNA translation). Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to fo ...
s appears to be a general mechanism for homo-oligomer (multimer) formation.Bertolini M, Fenzl K, Kats I, Wruck F, Tippmann F, Schmitt J, Auburger JJ, Tans S, Bukau B, Kramer G. Interactions between nascent proteins translated by adjacent ribosomes drive homomer assembly. Science. 2021 Jan 1;371(6524):57-64. . PMID 33384371 Hundreds of protein oligomers were identified that assemble in human cells by such an interaction. The most prevalent form of interaction is between the N-terminal regions of the interacting proteins. Dimer formation appears to be able to occur independently of dedicated assembly machines. The intermolecular forces likely responsible for self-recognition and multimer formation were discussed by Jehle.
Other potential methods
Diverse techniques to identify PPIs have been emerging along with technology progression. These include co-immunoprecipitation, protein microarrays,analytical ultracentrifugation Analytical ultracentrifugation is an analytical technique which combines an ultracentrifuge with optical monitoring systems.
In an analytical ultracentrifuge (commonly abbreviated as AUC), a sample’s sedimentation profile is monitored in real tim ...
, light scattering
Scattering is a term used in physics to describe a wide range of physical processes where moving particles or radiation of some form, such as light or sound, are forced to deviate from a straight trajectory by localized non-uniformities (including ...
, fluorescence spectroscopy
Fluorescence spectroscopy (also known as fluorimetry or spectrofluorometry) is a type of electromagnetic spectroscopy that analyzes fluorescence from a sample. It involves using a beam of light, usually ultraviolet light, that excites the electro ...
, luminescence-based mammalian interactome mapping (LUMIER), resonance-energy transfer systems, mammalian protein–protein interaction trap, electro-switchable biosurfaces, protein-fragment complementation assay Within the field of molecular biology, a protein-fragment complementation assay, or PCA, is a method for the identification and quantification of protein–protein interactions. In the PCA, the proteins of interest ("bait" and "prey") are each cov ...
, as well as real-time label-free measurements by surface plasmon resonance, and calorimetry
In chemistry and thermodynamics, calorimetry () is the science or act of measuring changes in ''state variables'' of a body for the purpose of deriving the heat transfer associated with changes of its state due, for example, to chemical re ...
.
Computational methods

Computational prediction of protein–protein interactions
The experimental detection and characterization of PPIs is labor-intensive and time-consuming. However, many PPIs can be also predicted computationally, usually using experimental data as a starting point. However, methods have also been developed that allow the prediction of PPI de novo, that is without prior evidence for these interactions.Genomic context methods
'' The Rosetta Stone or Domain Fusion method'' is based on the hypothesis that interacting proteins are sometimes fused into a single protein in another genome. Therefore, we can predict if two proteins may be interacting by determining if they each have non-overlapping sequence similarity to a region of a single protein sequence in another genome. '' The Conserved Neighborhood method'' is based on the hypothesis that if genes encoding two proteins are neighbors on a chromosome in many genomes, then they are likely functionally related (and possibly physically interacting)''.'' '' The Phylogenetic Profile method'' is based on the hypothesis that if two or more proteins are concurrently present or absent across several genomes, then they are likely functionally related. Therefore, potentially interacting proteins can be identified by determining the presence or absence of genes across many genomes and selecting those genes which are always present or absent together.Text mining methods
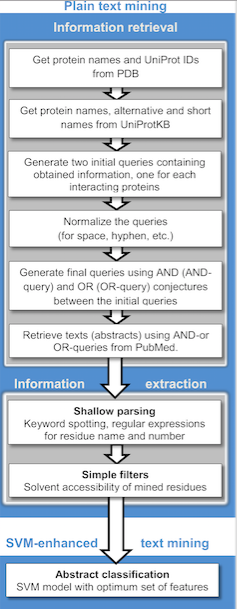
Publicly available information from biomedical documents is readily accessible through the internet and is becoming a powerful resource for collecting known protein-protein interactions (PPIs), PPI prediction and protein docking. Text mining is much less costly and time-consuming compared to other high-throughput techniques. Currently, text mining methods generally detectbinary relation
In mathematics, a binary relation associates elements of one set, called the ''domain'', with elements of another set, called the ''codomain''. A binary relation over sets and is a new set of ordered pairs consisting of elements in and in ...
s between interacting proteins from individual sentences using rule/pattern-based information extraction and machine learning
Machine learning (ML) is a field of inquiry devoted to understanding and building methods that 'learn', that is, methods that leverage data to improve performance on some set of tasks. It is seen as a part of artificial intelligence.
Machine ...
approaches. A wide variety of text mining applications for PPI extraction and/or prediction are available for public use, as well as repositories which often store manually validated and/or computationally predicted PPIs. Text mining can be implemented in two stages: ''information retrieval'', where texts containing names of either or both interacting proteins are retrieved and ''information extraction,'' where targeted information (interacting proteins, implicated residues, interaction types, etc.) is extracted.
There are also studies using phylogenetic profiling Phylogenetic profiling is a bioinformatics technique in which the joint presence or joint absence of two traits across large numbers of species is used to infer a meaningful biological connection, such as involvement of two different proteins in the ...
, basing their functionalities on the theory that proteins involved in common pathways co-evolve in a correlated fashion across species. Some more complex text mining methodologies use advanced Natural Language Processing
Natural language processing (NLP) is an interdisciplinary subfield of linguistics, computer science, and artificial intelligence concerned with the interactions between computers and human language, in particular how to program computers to proc ...
(NLP) techniques and build knowledge networks (for example, considering gene names as nodes and verbs as edges). Other developments involve kernel methods to predict protein interactions.
Machine learning methods
 Many computational methods have been suggested and reviewed for predicting protein-protein interactions. Prediction approaches can be grouped into categories based on predictive evidence: protein sequence,
Many computational methods have been suggested and reviewed for predicting protein-protein interactions. Prediction approaches can be grouped into categories based on predictive evidence: protein sequence, comparative genomics
Comparative genomics is a field of biological research in which the genomic features of different organisms are compared. The genomic features may include the DNA sequence, genes, gene order, regulatory sequences, and other genomic structural ...
, protein domains, protein tertiary structure, and interaction network topology. The construction of a positive set (known interacting protein pairs) and a negative set (non-interacting protein pairs) is needed for the development of a computational prediction model. Prediction models using machine learning techniques can be broadly classified into two main groups: supervised and unsupervised, based on the labeling of input variables according to the expected outcome.
In 2005, integral membrane proteins of Saccharomyces cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungus microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have b ...
were analyzed using the mating-based ubiquitin system (mbSUS). The system detects membrane proteins interactions with extracellular signaling proteins Of the 705 integral membrane proteins 1,985 different interactions were traced that involved 536 proteins. To sort and classify interactions a support vector machine was used to define high medium and low confidence interactions. The split-ubiquitin membrane yeast two-hybrid system uses transcriptional reporters to identify yeast transformants that encode pairs of interacting proteins.
In 2006, random forest
Random forests or random decision forests is an ensemble learning method for classification, regression and other tasks that operates by constructing a multitude of decision trees at training time. For classification tasks, the output of ...
, an example of a supervised technique, was found to be the most-effective machine learning method for protein interaction prediction. Such methods have been applied for discovering protein interactions on human interactome, specifically the interactome of Membrane proteins
Membrane proteins are common proteins that are part of, or interact with, biological membranes. Membrane proteins fall into several broad categories depending on their location. Integral membrane proteins are a permanent part of a cell membrane ...
and the interactome of Schizophrenia-associated proteins.
As of 2020, a model using residue cluster classes (RCCs), constructed from the 3DID and Negatome databases, resulted in 96-99% correctly classified instances of protein-protein interactions. RCCs are a computational vector space that mimics protein fold space and includes all simultaneously contacted residue sets, which can be used to analyze protein structure-function relation and evolution.
Databases
Large scale identification of PPIs generated hundreds of thousands of interactions, which were collected together in specializedbiological databases
Biological databases are libraries of biological sciences, collected from scientific experiments, published literature, high-throughput experiment technology, and computational analysis. They contain information from research areas including genom ...
that are continuously updated in order to provide complete interactomes. The first of these databases was the Database of Interacting Proteins (DIP).
''Primary databases'' collect information about published PPIs proven to exist via small-scale or large-scale experimental methods. Examples: DIP, Biomolecular Interaction Network Database (BIND), Biological General Repository for Interaction Datasets ( BioGRID), Human Protein Reference Database (HPRD), IntAct Molecular Interaction Database, Molecular Interactions Database (MINT), MIPS Protein Interaction Resource on Yeast (MIPS-MPact), and MIPS Mammalian Protein–Protein Interaction Database (MIPS-MPPI).<
''Meta-databases'' normally result from the integration of primary databases information, but can also collect some original data.
''Prediction databases'' include many PPIs that are predicted using several techniques (main article). Examples: Human Protein–Protein Interaction Prediction Database (PIPs), Interlogous Interaction Database (I2D), Known and Predicted Protein–Protein Interactions (STRING-db), and Unified Human Interactive (UniHI).
The aforementioned computational methods all depend on source databases whose data can be extrapolated to predict novel protein-protein interactions''. Coverage'' differs greatly between databases. In general, primary databases have the fewest total protein interactions recorded as they do not integrate data from multiple other databases, while prediction databases have the most because they include other forms of evidence in addition to experimental. For example, the primary database IntAct has 572,063 interactions, the meta-database APID has 678,000 interactions, and the predictive database STRING has 25,914,693 interactions. However, it is important to note that some of the interactions in the STRING database are only predicted by computational methods such as Genomic Context and not experimentally verified.
Interaction networks
 Information found in PPIs databases supports the construction of interaction networks. Although the PPI network of a given query protein can be represented in textbooks, diagrams of whole cell PPIs are frankly complex and difficult to generate.
One example of a manually produced molecular interaction map is the Kurt Kohn's 1999 map of cell cycle control. Drawing on Kohn's map, Schwikowski et al. in 2000 published a paper on PPIs in yeast, linking 1,548 interacting proteins determined by two-hybrid screening. They used a layered graph drawing method to find an initial placement of the nodes and then improved the layout using a force-based algorithm.
Bioinformatic tools have been developed to simplify the difficult task of visualizing molecular interaction networks and complement them with other types of data. For instance,
Information found in PPIs databases supports the construction of interaction networks. Although the PPI network of a given query protein can be represented in textbooks, diagrams of whole cell PPIs are frankly complex and difficult to generate.
One example of a manually produced molecular interaction map is the Kurt Kohn's 1999 map of cell cycle control. Drawing on Kohn's map, Schwikowski et al. in 2000 published a paper on PPIs in yeast, linking 1,548 interacting proteins determined by two-hybrid screening. They used a layered graph drawing method to find an initial placement of the nodes and then improved the layout using a force-based algorithm.
Bioinformatic tools have been developed to simplify the difficult task of visualizing molecular interaction networks and complement them with other types of data. For instance, Cytoscape
Cytoscape is an open source bioinformatics software platform for visualizing molecular interaction networks and integrating with gene expression profiles and other state data. Additional features are available as plugins. Plugins are available fo ...
is an open-source software widely used and many plugins are currently available. Pajek software is advantageous for the visualization and analysis of very large networks.
Identification of functional modules in PPI networks is an important challenge in bioinformatics. Functional modules means a set of proteins that are highly connected to each other in PPI network. It is almost similar problem as community detection in social network
A social network is a social structure made up of a set of social actors (such as individuals or organizations), sets of dyadic ties, and other social interactions between actors. The social network perspective provides a set of methods fo ...
s. There are some methods such as Jactive modules and MoBaS. Jactive modules integrate PPI network and gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. T ...
data where as MoBaS integrate PPI network and Genome Wide association Studies.
Protein-protein relationships are often the result of multiple types of interactions or are deduced from different approaches, including co-localization, direct interaction, suppressive genetic interaction, additive genetic interaction, physical association, and other associations.
Signed interaction networks
 Protein–protein interactions often result in one of the interacting proteins either being 'activated' or 'repressed'. Such effects can be indicated in a PPI network by "signs" (e.g. "activation" or "inhibition"). Although such attributes have been added to networks for a long time, Vinayagam et al. (2014) coined the term ''Signed network'' for them. Signed networks are often expressed by labeling the interaction as either positive or negative. A positive interaction is one where the interaction results in one of the proteins being activated. Conversely, a negative interaction indicates that one of the proteins being inactivated.
Protein–protein interaction networks are often constructed as a result of lab experiments such as yeast two-hybrid screens or 'affinity purification and subsequent mass spectrometry techniques. However these methods do not provide the layer of information needed in order to determine what type of interaction is present in order to be able to attribute signs to the network diagrams.
Protein–protein interactions often result in one of the interacting proteins either being 'activated' or 'repressed'. Such effects can be indicated in a PPI network by "signs" (e.g. "activation" or "inhibition"). Although such attributes have been added to networks for a long time, Vinayagam et al. (2014) coined the term ''Signed network'' for them. Signed networks are often expressed by labeling the interaction as either positive or negative. A positive interaction is one where the interaction results in one of the proteins being activated. Conversely, a negative interaction indicates that one of the proteins being inactivated.
Protein–protein interaction networks are often constructed as a result of lab experiments such as yeast two-hybrid screens or 'affinity purification and subsequent mass spectrometry techniques. However these methods do not provide the layer of information needed in order to determine what type of interaction is present in order to be able to attribute signs to the network diagrams.
RNA interference screens
RNA interference
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by ...
(RNAi) screens (repression of individual proteins between transcription and translation) are one method that can be utilized in the process of providing signs to the protein-protein interactions. Individual proteins are repressed and the resulting phenotypes are analyzed. A correlating phenotypic relationship (i.e. where the inhibition of either of two proteins results in the same phenotype) indicates a positive, or activating relationship. Phenotypes that do not correlate (i.e. where the inhibition of either of two proteins results in two different phenotypes) indicate a negative or inactivating relationship. If protein A is dependent on protein B for activation then the inhibition of either protein A or B will result in a cell losing the service that is provided by protein A and the phenotypes will be the same for the inhibition of either A or B. If, however, protein A is inactivated by protein B then the phenotypes will differ depending on which protein is inhibited (inhibit protein B and it can no longer inactivate protein A leaving A active however inactivate A and there is nothing for B to activate since A is inactive and the phenotype changes). Multiple RNAi screens need to be performed in order to reliably appoint a sign to a given protein-protein interaction. Vinayagam et al. who devised this technique state that a minimum of nine RNAi screens are required with confidence increasing as one carries out more screens.
As therapeutic targets
Modulation of PPI is challenging and is receiving increasing attention by the scientific community. Several properties of PPI such as allosteric sites and hotspots, have been incorporated into drug-design strategies. The relevance of PPI as putative therapeutic targets for the development of new treatments is particularly evident in cancer, with several ongoing clinical trials within this area. The consensus among these promising targets is, nonetheless, denoted in the already available drugs on the market to treat a multitude of diseases. Examples are Tirobifan, inhibitor of the glycoprotein IIb/IIIa, used as a cardiovascular drug, and Maraviroc, inhibitor of the CCR5-gp120 interaction, used as anti-HIV drug. Recently, Amit Jaiswal and others were able to develop 30 peptides using protein–protein interaction studies to inhibit telomerase recruitment towards telomeres.See also
*Glycan-protein interactions
Glycan-Protein interactions represent a class of biomolecular interactions that occur between free or protein-bound glycans and their cognate binding partners. Intramolecular glycan-protein (protein-glycan) interactions occur between glycans and p ...
* 3did
*Allostery
In biochemistry, allosteric regulation (or allosteric control) is the regulation of an enzyme by binding an effector molecule at a site other than the enzyme's active site.
The site to which the effector binds is termed the ''allosteric site ...
* Biological network
* Biological machines
*DIMA (database)
The Domain Interaction MAp (DIMA) is a database of predicted and known interactions between protein domains. Version 3.0 of the database was released in 2010.
See also
* Protein domain
In molecular biology, a protein domain is a region of a ...
*Enzyme catalysis
Enzyme catalysis is the increase in the rate of a process by a biological molecule, an " enzyme". Most enzymes are proteins, and most such processes are chemical reactions. Within the enzyme, generally catalysis occurs at a localized site, cal ...
* HitPredict
* Human interactome
* IsoBase
*Multiprotein complex
A protein complex or multiprotein complex is a group of two or more associated polypeptide chains. Protein complexes are distinct from multienzyme complexes, in which multiple catalytic domains are found in a single polypeptide chain.
Protein ...
* Protein domain dynamics
* Protein flexibility
*Protein structure
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, the monomers of the polymer. A single amino acid monom ...
* Protein–protein interaction prediction
* Protein–protein interaction screening
*Systems biology
Systems biology is the computational and mathematical analysis and modeling of complex biological systems. It is a biology-based interdisciplinary field of study that focuses on complex interactions within biological systems, using a holistic ...
References
Further reading
* * * * * *External links
Protein-Protein Interaction Databases
Library of Modulators of Protein–Protein Interactions (PPI)
* * {{DEFAULTSORT:Protein-Protein Interaction Proteomics Signal transduction Biophysics Biochemistry methods Biotechnology Quantum biochemistry Protein-protein interaction assays Protein complexes