|
Protein–protein Interaction Screening
Protein–protein interaction screening refers to the identification of Protein–protein interaction with high-throughput screening methods such as computer- and/or robot-assisted plate reading, flow cytometry analyzing. The interactions between proteins are central to virtually every process in a living cell. Information about these interactions improves understanding of diseases and can provide the basis for new therapeutic approaches. Methods to screen protein–protein interactions Though there are many methods to detect protein–protein interactions, the majority of these methods—such as co-immunoprecipitation, fluorescence resonance energy transfer (FRET) and dual polarisation interferometry—are not screening approaches. ''Ex vivo'' or ''in vivo'' methods Methods that screen protein–protein interactions in the living cells. Bimolecular fluorescence complementation (BiFC) is a technique for observing the interactions of proteins. Combining it with other new techniqu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein–protein Interaction
Protein–protein interactions (PPIs) are physical contacts of high specificity established between two or more protein molecules as a result of biochemical events steered by interactions that include electrostatic forces, hydrogen bonding and the hydrophobic effect. Many are physical contacts with molecular associations between chains that occur in a cell or in a living organism in a specific biomolecular context. Proteins rarely act alone as their functions tend to be regulated. Many molecular processes within a cell are carried out by molecular machines that are built from numerous protein components organized by their PPIs. These physiological interactions make up the so-called interactomics of the organism, while aberrant PPIs are the basis of multiple aggregation-related diseases, such as Creutzfeldt–Jakob and Alzheimer's diseases. PPIs have been studied with many methods and from different perspectives: biochemistry, quantum chemistry, molecular dynamics, signal trans ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
High-throughput Screening
High-throughput screening (HTS) is a method for scientific experimentation especially used in drug discovery and relevant to the fields of biology, materials science and chemistry. Using robotics, data processing/control software, liquid handling devices, and sensitive detectors, high-throughput screening allows a researcher to quickly conduct millions of chemical, genetic, or pharmacological tests. Through this process one can quickly recognize active compounds, antibodies, or genes that modulate a particular biomolecular pathway. The results of these experiments provide starting points for drug design and for understanding the noninteraction or role of a particular location. Assay plate preparation The key labware or testing vessel of HTS is the microtiter plate, which is a small container, usually disposable and made of plastic, that features a grid of small, open divots called ''wells''. In general, microplates for HTS have either 96, 192, 384, 1536, 3456 or 6144 wells. The ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Co-immunoprecipitation
Immunoprecipitation (IP) is the technique of precipitating a protein antigen out of solution using an antibody that specifically binds to that particular protein. This process can be used to isolate and concentrate a particular protein from a sample containing many thousands of different proteins. Immunoprecipitation requires that the antibody be coupled to a solid substrate at some point in the procedure. Types Individual protein immunoprecipitation (IP) Involves using an antibody that is specific for a known protein to isolate that particular protein out of a solution containing many different proteins. These solutions will often be in the form of a crude lysate of a plant or animal tissue. Other sample types could be body fluids or other samples of biological origin. Protein complex immunoprecipitation (Co-IP) Immunoprecipitation of intact protein complexes (i.e. antigen along with any proteins or ligands that are bound to it) is known as co-immunoprecipitation (Co-I ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fluorescence Resonance Energy Transfer
Fluorescence is the emission of light by a substance that has absorbed light or other electromagnetic radiation. It is a form of luminescence. In most cases, the emitted light has a longer wavelength, and therefore a lower photon energy, than the absorbed radiation. A perceptible example of fluorescence occurs when the absorbed radiation is in the ultraviolet region of the electromagnetic spectrum (invisible to the human eye), while the emitted light is in the visible region; this gives the fluorescent substance a distinct color that can only be seen when the substance has been exposed to UV light. Fluorescent materials cease to glow nearly immediately when the radiation source stops, unlike phosphorescent materials, which continue to emit light for some time after. Fluorescence has many practical applications, including mineralogy, gemology, medicine, chemical sensors (fluorescence spectroscopy), fluorescent labelling, dyes, biological detectors, cosmic-ray detection, vacu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dual Polarisation Interferometry
Dual-polarization interferometry (DPI) is an analytical technique that probes molecular layers adsorbed to the surface of a waveguide using the evanescent wave of a laser beam. It is used to measure the conformational change in proteins, or other biomolecules, as they function (referred to as the conformation activity relationship). Instrumentation DPI focuses laser light into two waveguides. One of these functions as the "sensing" waveguide having an exposed surface while the second one functions to maintain a reference beam. A two-dimensional interference pattern is formed in the far field by combining the light passing through the two waveguides. The DPI technique rotates the polarization of the laser, to alternately excite two polarization modes of the waveguides. Measurement of the interferogram for both polarizations allows both the refractive index and the thickness of the adsorbed layer to be calculated. The polarization can be switched rapidly, allowing real-time measur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
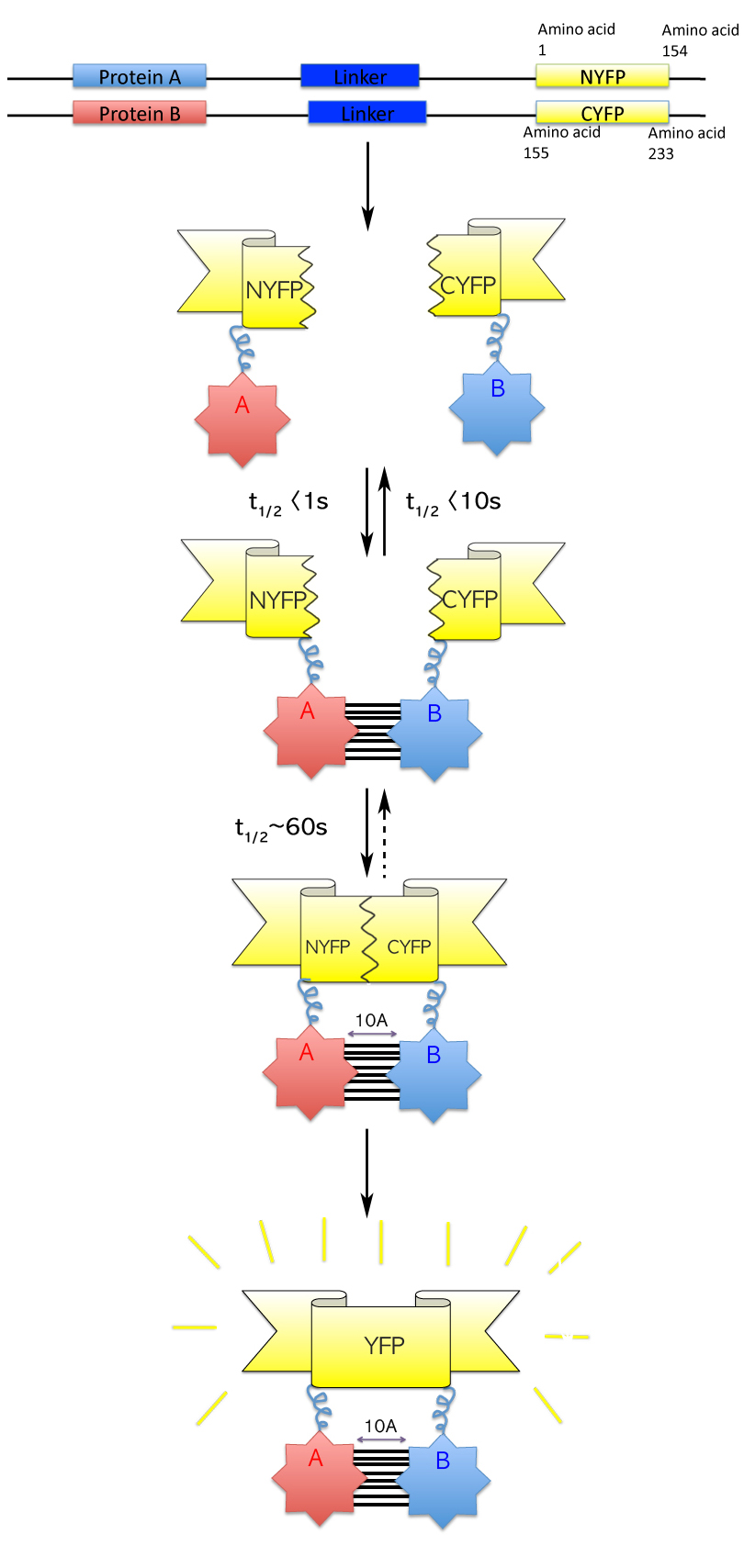
Bimolecular Fluorescence Complementation
Bimolecular fluorescence complementation (also known as BiFC) is a technology typically used to validate protein interactions. It is based on the association of fluorescent protein fragments that are attached to components of the same macromolecular complex. Proteins that are postulated to interact are fused to unfolded complementary fragments of a fluorescent reporter protein and expressed in live cells. Interaction of these proteins will bring the fluorescent fragments within proximity, allowing the reporter protein to reform in its native three-dimensional structure and emit its fluorescent signal.Kerppola, T. K. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 1, 1278–1286 (2006). This fluorescent signal can be detected and located within the cell using an inverted fluorescence microscope that allows imaging of fluorescence in cells. In addition, the intensity ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nature Precedings
''Nature Precedings'' was an open access electronic preprint repository of scholarly work in the fields of biomedical sciences, chemistry, and earth sciences. It ceased accepting new submissions as of April 3, 2012. ''Nature Precedings'' functioned as a permanent, citable archive for pre-publication research and preliminary findings. It was a place for researchers to share documents, including presentations, posters, white papers, technical papers, supplementary findings, and non-peer-reviewed manuscripts. It provided a rapid way to share preliminary findings, disseminate emerging results, solicit community feedback, and claim priority over discoveries. The content was curated and developed by the Nature Publishing Group. Description ''Nature Precedings'' was started in June 2007 by the Nature Publishing Group under the direction of Timo Hannay, its director for web publishing. The British Library, the European Bioinformatics Institute, Science Commons, and the Wellcome Trust wer ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Yeast Two-hybrid
Two-hybrid screening (originally known as yeast two-hybrid system or Y2H) is a molecular biology technique used to discover protein–protein interactions (PPIs) and protein–DNA interactions by testing for physical interactions (such as binding) between two proteins or a single protein and a DNA molecule, respectively. The premise behind the test is the activation of downstream reporter gene(s) by the binding of a transcription factor onto an upstream activating sequence (UAS). For two-hybrid screening, the transcription factor is split into two separate fragments, called the DNA-binding domain (DBD or often also abbreviated as BD) and activating domain (AD). The BD is the domain responsible for binding to the UAS and the AD is the domain responsible for the activation of transcription. The Y2H is thus a protein-fragment complementation assay. History Pioneered by Stanley Fields and Ok-Kyu Song in 1989, the technique was originally designed to detect protein–protein ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FEBS Journal
''The FEBS Journal'' is a biweekly peer-reviewed scientific journal published by John Wiley & Sons on behalf of the Federation of European Biochemical Societies. It covers research on all aspects of biochemistry, molecular biology, cell biology, and the molecular bases of disease. The editor-in-chief is Seamus Martin (Trinity College Dublin), who took over from Richard Perham (University of Cambridge) in 2014. Content is available for free 1 year after publication, except review content, which is available immediately. The journal also publishes special and virtual issues focusing on a specific theme. Since 2021, the journal has given an annual award, "The FEBS Journal Richard Perham Prize", for an outstanding research paper published in the journal. The winners receive a €5,000 cash prize (to be divided equally between the first and last authors) and the senior author of the study is invited to give a talk at the FEBS Annual Congress. The journal also gives more frequent post ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tandem Affinity Purification
Tandem affinity purification (TAP) is an immunoprecipitation-based purification technique for studying protein–protein interactions. The goal is to extract from a cell only the protein of interest, in complex with any other proteins it interacted with. TAP uses two types of agarose beads that bind to the protein of interest and that can be separated from the cell lysate by centrifugation, without disturbing, denaturing or contaminating the involved complexes. To enable the protein of interest to bind to the beads, it is tagged with a designed piece, the TAP tag. The original TAP method involves the fusion of the TAP tag to the C-terminus of the protein under study. The TAP tag consists of three components: a calmodulin binding peptide (CBP), TEV protease cleavage site, and two Protein A domains, which bind tightly to IgG (making a TAP tag a type of epitope tag). Many other tag/bead/eluent combinations have been proposed since the TAP principle was first published. Variant ta ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cross-link
In chemistry and biology a cross-link is a bond or a short sequence of bonds that links one polymer chain to another. These links may take the form of covalent bonds or ionic bonds and the polymers can be either synthetic polymers or natural polymers (such as proteins). In polymer chemistry "cross-linking" usually refers to the use of cross-links to promote a change in the polymers' physical properties. When "crosslinking" is used in the biological field, it refers to the use of a probe to link proteins together to check for protein–protein interactions, as well as other creative cross-linking methodologies. Although the term is used to refer to the "linking of polymer chains" for both sciences, the extent of crosslinking and specificities of the crosslinking agents vary greatly. As with all science, there are overlaps, and the following delineations are a starting point to understanding the subtleties. Polymer chemistry Crosslinking is the general term for the process of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |