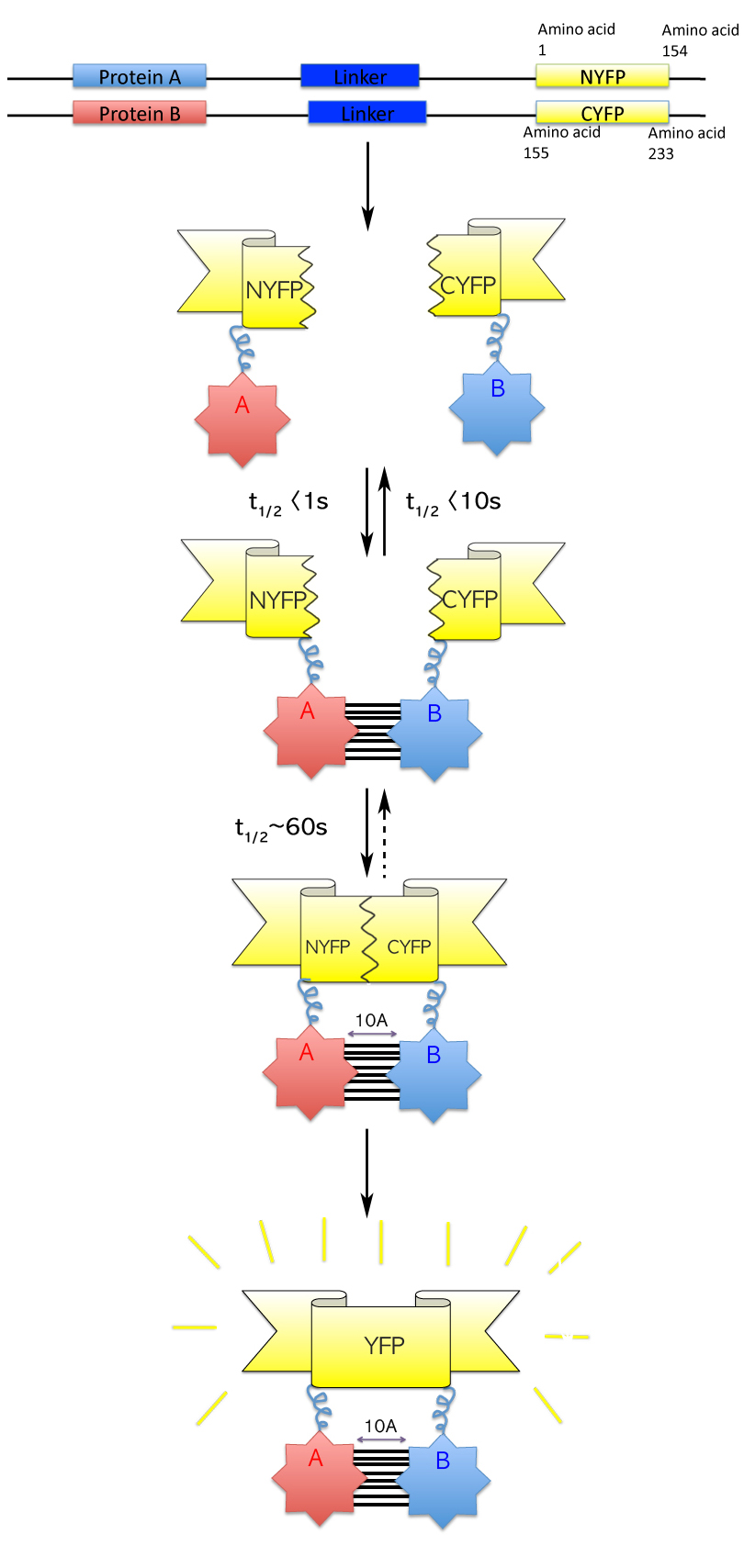
Bimolecular fluorescence complementation (also known as BiFC) is a technology typically used to validate
protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, respo ...
interactions. It is based on the association of fluorescent protein fragments that are attached to components of the same
macromolecular
A macromolecule is a very large molecule important to biophysical processes, such as a protein or nucleic acid. It is composed of thousands of covalently bonded atoms. Many macromolecules are polymers of smaller molecules called monomers. The ...
complex. Proteins that are postulated to interact are fused to unfolded complementary fragments of a fluorescent
reporter protein and expressed in live cells. Interaction of these proteins will bring the fluorescent fragments within proximity, allowing the reporter protein to reform in its
native three-dimensional structure and emit its fluorescent signal.
[Kerppola, T. K. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 1, 1278–1286 (2006).] This fluorescent signal can be detected and located within the cell using an
inverted fluorescence microscope that allows imaging of fluorescence in cells. In addition, the intensity of the fluorescence emitted is proportional to the strength of the interaction, with stronger levels of fluorescence indicating close or direct interactions and lower fluorescence levels suggesting interaction within a complex.
[Morell, M., Espargaro, A., Aviles, F. X. & Ventura, S. Study and selection of in vivo protein interactions by coupling bimolecular fluorescence complementation and flow cytometry. Nat. Protoc. 3, 22–33 (2008).] Therefore, through the visualisation and analysis of the intensity and distribution of fluorescence in these cells, one can identify both the location and interaction partners of proteins of interest.
History
Biochemical
complementation was first reported in subtilisin-cleaved
bovine
Bovines (subfamily Bovinae) comprise a diverse group of 10 genera of medium to large-sized ungulates, including cattle, bison, African buffalo, water buffalos, and the four-horned and spiral-horned antelopes. The evolutionary relationship betwee ...
pancreatic
ribonuclease
Ribonuclease (commonly abbreviated RNase) is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the ...
, then expanded using
β-galactosidase
β-Galactosidase (EC 3.2.1.23, lactase, beta-gal or β-gal; systematic name β-D-galactoside galactohydrolase), is a glycoside hydrolase enzyme that catalyst, catalyzes hydrolysis of terminal non-reducing β-D-galactose residues in β-D-galactosi ...
mutants that allowed cells to grow on lactose.
[Richards, F. M. On the Enzymic Activity of Subtilisin-Modified Ribonuclease. Proc. Natl. Acad. Sci. U. S. A. 44, 162–166 (1958).][Ullmann, A., Jacob, F. & Monod, J. Characterization by in vitro complementation of a peptide corresponding to an operator-proximal segment of the beta-galactosidase structural gene of Escherichia coli. J. Mol. Biol. 24, 339–343 (1967).][Ullmann, A., Jacob, F. & Monod, J. On the subunit structure of wild-type versus complemented beta-galactosidase of Escherichia coli. J. Mol. Biol. 32, 1–13 (1968).]
Recognition of many proteins' ability to spontaneously assemble into functional complexes as well as the ability of protein fragments to assemble as a consequence of the spontaneous functional complex assembly of interaction partners to which they are fused was later reported for
ubiquitin
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Fo ...
fragments in yeast protein interactions.
[Johnsson, N. & Varshavsky, A. Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. U. S. A. 91, 10340-10344 (1994).]
In 2000, Ghosh ''et al'' developed a system that allowed a
green fluorescent protein
The green fluorescent protein (GFP) is a protein that exhibits bright green fluorescence when exposed to light in the blue to ultraviolet range. The label ''GFP'' traditionally refers to the protein first isolated from the jellyfish ''Aequorea ...
(
GFP GFP may refer to:
Organisations
* Gaelic Football Provence, a French Gaelic Athletic Association club
* Geheime Feldpolizei, the German secret military police during the Second World War
* French Group for the Study of Polymers and their Applicat ...
) to be reassembled using an
anti-parallel leucine zipper
A leucine zipper (or leucine scissors) is a common three-dimensional structural motif in proteins. They were first described by Landschulz and collaborators in 1988 when they found that an enhancer binding protein had a very characteristic 30-amin ...
in ''E. coli'' cells.
[Ghosh, I., Hamilton, A. D. & Regan, L. Antiparallel Leucine Zipper-Directed Protein Reassembly: Application to the Green Fluorescent Protein. Journal of the American Chemical Society 122, 5658 (2000).] This was achieved by dissecting GFP into
C- and
N-terminal
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the ami ...
GFP fragments. As the GFP fragment was attached to each leucine zipper by a linker, the heterodimerisation of the anti-parallel leucine zipper resulted in a reconstituted, or re-formed, GFP protein that could be visualised. The successful fluorescent signal indicated that the separate GFP peptide fragments were able to correctly reassemble and achieve
tertiary folding. It was, therefore, postulated that using this technique, fragmented GFP could be used to study
interaction of protein–protein pairs that have their N–C termini in close proximity.
After the demonstration of successful fluorescent protein fragment reconstitution in mammalian cells, Hu ''et al''. described the use of fragmented
yellow fluorescent protein
Yellow fluorescent protein (YFP) is a genetic mutant of green fluorescent protein (GFP) originally derived from the jellyfish ''Aequorea victoria''. Its excitation peak is 513 nm and its emission peak is 527 nm. Like the parent GFP, YFP ...
(
YFP) in the investigation of bZIP and Rel family
transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The fu ...
interactions.
This was the first report bZIP protein interaction regulation by regions outside of the
bZIP domain
The Basic Leucine Zipper Domain (bZIP domain) is found in many DNA binding eukaryotic proteins. One part of the domain contains a region that mediates sequence specific DNA binding properties and the leucine zipper that is required to hold tog ...
, regulation of
subnuclear localization of the bZIP domains Fos and
Jun by their different interacting partners, and modulation of
transcriptional activation of bZIP and Rel proteins through mutual interactions. In addition, this study was the first report of an ''
in vivo
Studies that are ''in vivo'' (Latin for "within the living"; often not italicized in English) are those in which the effects of various biological entities are tested on whole, living organisms or cells, usually animals, including humans, and ...
'' technique, now known as the bimolecular fluorescence complementation (BiFC) assay, to provide insight into the structural basis of protein complex formation through detection of fluorescence caused by the assembly of fluorescent reporter protein fragments tethered to interacting proteins.
[Hu, C. D., Chinenov, Y. & Kerppola, T. K. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9, 789–798 (2002).]
Fluorescent labeling
Fluorophore
A fluorophore (or fluorochrome, similarly to a chromophore) is a fluorescent chemical compound that can re-emit light upon light excitation. Fluorophores typically contain several combined aromatic groups, or planar or cyclic molecules with se ...
activation occurs through an autocatalytic cyclization reaction that occurs after the protein has been folded correctly.
[Tsien, R. Y. The green fluorescent protein. Annu. Rev. Biochem. 67, 509–544 (1998).] This was advanced with the successful reconstitution of the YFP fluorophore from protein fragments that had been fused to interacting proteins within 8 hours of transfection, reported in 2002.
Workflow

Selection of fusion protein production system
There are different
production systems that can be used for the fusion protein generated. Transient gene expression is used to identify protein–protein interactions ''in vivo'' as well as in subcellular localisation of the BiFC complex. However, one must be cautious against protein over-expression, as this may skew both preferential localisation and the predominant protein complexes formed. Instead, weak
promoters, the use of low levels of plasmid DNA in the transfection, and plasmid vectors that do not replicate in mammalian cells should be used to express proteins at or near their endogenous levels to mimic the physiological cellular environment.
Also, careful selection of the fluorescent protein is important, as different fluorescent proteins require different cellular environments. For example, GFP can be used in ''E. coli'' cells, while YFP is used in mammalian cells.
[Kerppola, T. K. Complementary methods for studies of protein interactions in living cells. Nat. Methods 3, 969–971 (2006).]
Stable
cell line
An immortalised cell line is a population of cells from a multicellular organism which would normally not proliferate indefinitely but, due to mutation, have evaded normal cellular senescence and instead can keep undergoing division. The cell ...
s with the expression vector integrated into its genome allows more stable
gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. The ...
in the cell population, resulting in more consistent results.
Determination of fusion sites
When deciding the linker fusion site on the protein surface, there are three main considerations. First, the fluorescent protein fragments must be able to associate with one another when their tethered proteins interact.
Structural information and the location of the interaction surface may be useful when determining the fusion site to the linker, although the information is not necessary, as multiple combinations and permutations can be screened.
Secondly, the creation of the fusion protein must not significantly alter the localisation, stability, or expression of the proteins to which the fragments are linked as compared to the
endogenous
Endogenous substances and processes are those that originate from within a living system such as an organism, tissue, or cell.
In contrast, exogenous substances and processes are those that originate from outside of an organism.
For example, es ...
wild-type
The wild type (WT) is the phenotype of the typical form of a species as it occurs in nature. Originally, the wild type was conceptualized as a product of the standard "normal" allele at a locus, in contrast to that produced by a non-standard, "m ...
proteins.
Finally, the addition of the fluorescent fragment fusion must not affect the biological function of the protein, preferably verified using assays that evaluate all of the proteins' known functions.
Designing linkers
A linker is a short
amino acid sequence
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthes ...
that tethers the fluorescent reporter protein fragment to the protein of interest, forming the fusion protein. When designing a linker sequence, one must ensure that the linker is sufficiently
soluble
In chemistry, solubility is the ability of a substance, the solute, to form a solution with another substance, the solvent. Insolubility is the opposite property, the inability of the solute to form such a solution.
The extent of the solubil ...
and long to provide the fluorescent protein fragments with flexibility and freedom of movement so that the fragment and its partner fragment will collide frequently enough to reconstitute during the interaction of their respective fused proteins.
Although it is not documented, it is possible that the length or the sequence of the linker may influence complementation of some proteins.
Reported linker sequences RSIAT and RPACKIPNDLKQKVMNH (single amino acid code) and AAANSSIDLISVPVDSR (Sigma) have been successfully used in BiFC experiments.
[Hu, C. D. & Kerppola, T. K. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21, 539–545 (2003).]
Creating proper plasmid expression vectors
When designing
plasmid vectors to express the proteins of interest, the
construct
Construct, Constructs or constructs may refer to:
* Construct (information technology), a collection of logic components forming an interactive agent or environment
** Language construct
* ''Construct'' (album), a 2013 album by Dark Tranquillity ...
must be able to express proteins that are able to form
fusion protein
Fusion proteins or chimeric (kī-ˈmir-ik) proteins (literally, made of parts from different sources) are proteins created through the joining of two or more genes that originally coded for separate proteins. Translation of this ''fusion gene'' r ...
s with fluorescent protein fragments without disrupting the protein's function. In addition, the expected protein complex must be able to accept stabilisation of the fluorescent protein fragment interaction without affecting the protein complex function or the cell being studied. Many fluorescent protein fragments that combine in several ways can be used in BiFC.
Generally, YFP is recommended to serve as the reporter protein, cleaved at
residue
Residue may refer to:
Chemistry and biology
* An amino acid, within a peptide chain
* Crop residue, materials left after agricultural processes
* Pesticide residue, refers to the pesticides that may remain on or in food after they are applied ...
155 (N-terminal consisting of residues 1–154 and C-terminal consisting of residues 155–238) or residue 173 in particular, as these sets of fragments are highly efficient in their complementation when fused to many interacting proteins and they produce low levels fluorescence when fused to non-interacting proteins. It is suggested that each target protein is fused to both the N- and C-terminal fragments of the fluorescent reporter protein in turn, and that the fragments are fused at each of the N- and C-terminal ends of the target proteins. This will allow a total of eight different permutations, with interactions being tested:
N-terminal fragment fused at the N-terminal protein 1 + C-terminal fragment fused at the N-terminal protein 2
N-terminal fragment fused at the N-terminal protein 1 + C-terminal fragment fused at the C-terminal protein 2
N-terminal fragment fused at the C-terminal protein 1 + C-terminal fragment fused at the N-terminal protein 2
N-terminal fragment fused at the C-terminal protein 1 + C-terminal fragment fused at the C-terminal protein 2
C-terminal fragment fused at the N-terminal protein 1 + N-terminal fragment fused at the N-terminal protein 2
C-terminal fragment fused at the N-terminal protein 1 + N-terminal fragment fused at the C-terminal protein 2
C-terminal fragment fused at the C-terminal protein 1 + N-terminal fragment fused at the N-terminal protein 2
C-terminal fragment fused at the C-terminal protein 1 + N-terminal fragment fused at the C-terminal protein 2
Selection of appropriate cell culture system
As previously stated, it is important to ensure that the fluorescent reporter protein being used in BiFC is appropriate and can be expressed in the
cell culture
Cell culture or tissue culture is the process by which cells are grown under controlled conditions, generally outside of their natural environment. The term "tissue culture" was coined by American pathologist Montrose Thomas Burrows. This te ...
system of choice, as not all reporter proteins can fluoresce or be visualised in all
model systems.
Selection of appropriate controls
Fluorescent protein fragments can associate and fluoresce at low efficiency in the absence of a specific interaction. Therefore, it is important to include
control
Control may refer to:
Basic meanings Economics and business
* Control (management), an element of management
* Control, an element of management accounting
* Comptroller (or controller), a senior financial officer in an organization
* Controllin ...
s to ensure that the fluorescence from fluorescent reporter protein reconstitution is not due to unspecific contact.
[Morell, M. et al. Monitoring the interference of protein–protein interactions in vivo by bimolecular fluorescence complementation: the DnaK case. Proteomics 8, 3433–3442 (2008).]
Some controls include fluorophore fragments linked to non-interacting proteins, as the presence of these fusions tend to decrease non-specific complementation and
false positive
A false positive is an error in binary classification in which a test result incorrectly indicates the presence of a condition (such as a disease when the disease is not present), while a false negative is the opposite error, where the test result ...
results.
Another control is created by linking the fluorescent protein fragment to proteins with mutated interaction faces.
So long as the fluorescent fragment is fused to the mutated proteins in the same manner as the wild-type protein, and the gene expression levels and localisation are unaffected by the
mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mi ...
, this serves as a strong negative control, as the mutant proteins, and therefore, the fluorescent fragments, should be unable to interact.
Internal controls are also necessary to normalise for differences in transfection efficiencies and gene expression in different cells. This is accomplished by co-transfecting cells with plasmids encoding the fusion proteins of interest as well as a whole (non-fragmented) protein that fluoresces at a different wavelength from the fluorescent reporter protein. During visualisation, one determines the fluorescence intensities of the BiFC complex and the internal control which, after subtracting background signal, becomes a ratio. This ratio represents the BiFC efficiency and can be compared with other ratios to determine the relative efficiencies of the formation of different complexes.
Cell transfection
Once the fusion proteins and controls have been designed and generated in their appropriate expression system, the plasmids must be
transfected
Transfection is the process of deliberately introducing naked or purified nucleic acids into eukaryotic cells. It may also refer to other methods and cell types, although other terms are often preferred: "transformation" is typically used to desc ...
into the cells to be studied. After transfection, one must wait, typically about eight hours, to allow time for the fusion proteins to interact and their linked fluorescent reporter protein fragments to associate and fluoresce.
Visualisation and analysis
After sufficient time for the fusion proteins and their linked fluorescent fragments to interact and fluoresce, the cells can be observed under an inverted fluorescence microscope that can visualise fluorescence in cells. Although the fluorescence intensity of BiFC complexes is usually <10% of that produced by expression of intact fluorescent proteins, the extremely low
autofluorescence
Autofluorescence is the natural emission of light by biological structures such as mitochondria and lysosomes when they have absorbed light, and is used to distinguish the light originating from artificially added fluorescent markers (fluorophores) ...
in the visible range extremely most cells often makes the BiFC signal orders of magnitude higher than background fluorescence.
[Kerppola, T. K. Bimolecular fluorescence complementation (BiFC) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37, 465–487 (2008).]
If fluorescence is detected when the fusion proteins are expressed, but is lacking or significantly reduced after the expression of the mutated negative control, it is likely that a specific interaction occurs between the two target proteins of interest. However, if the fluorescence intensity is not significantly different between the mutated negative control fusion protein and its wild-type counterpart, then the fluorescence is likely caused by non-specific protein interactions, so a different combination of fusion protein conformations should be tested.
If no fluorescence is detected, an interaction may still exist between the proteins of interest, as the creation of the fusion protein may alter the structure or interaction face of the target protein or the fluorescence fragments may be physically unable to associate. To ensure that this result is not a
false negative
A false positive is an error in binary classification in which a test result incorrectly indicates the presence of a condition (such as a disease when the disease is not present), while a false negative is the opposite error, where the test result ...
, that there is no interaction, the protein interaction must be tested in a situation where fluorescence complementation and activation requires an external signal. In this case, if the external signal fails to cause fluorescence fragment association, it is likely that the proteins do not interact or there is a physical impediment to fluorescence complementation.
Strengths
Relevant biological context
Proteins interact with different protein partners and other macromolecules to achieve functions that support different functions in cells that support survival of the organism. Identifying these interactions may provide clues to their effects on cell processes. As these interactions can be affected by both the internal environment and external stimuli, studying these interactions ''in vivo'' and at endogenous levels, as is recommended in BiFC, provides a physiologically-relevant context from which to draw conclusions about protein interactions.
Direct visualization
BiFC enables direct visualisation of protein interactions in living cells with limited cell
perturbation
Perturbation or perturb may refer to:
* Perturbation theory, mathematical methods that give approximate solutions to problems that cannot be solved exactly
* Perturbation (geology), changes in the nature of alluvial deposits over time
* Perturbatio ...
, rather than relying on secondary effects or staining by
exogenous
In a variety of contexts, exogeny or exogeneity () is the fact of an action or object originating externally. It contrasts with endogeneity or endogeny, the fact of being influenced within a system.
Economics
In an economic model, an exogeno ...
molecules that can fail to distribute evenly.
This, and the ability to observe the living cells for long periods of time, is made possible by the strong intrinsic fluorescence of the reconstituted reporter protein reduces the chances of an incorrect readout associated with the protein isolation process.
Sensitivity
Unlike many ''in vivo'' protein-interaction assays, BiFC does not require protein complexes to be formed by a large proportion of the proteins or at
stoichiometric
Stoichiometry refers to the relationship between the quantities of reactants and products before, during, and following chemical reactions.
Stoichiometry is founded on the law of conservation of mass where the total mass of the reactants equal ...
proportions. Instead, BiFC can detect interactions among protein
subpopulations, weak interactions, and low expression proteins due to the stable complementation of the fluorescent reporter protein.
In addition, successful fluorescent protein reconstitution has been reported for protein partners over 7 nm apart, so long as the linkers binding the fluorophore fragment to the protein of interest has the flexibility needed to associate with its corresponding fragment.
Furthermore, the strength of the protein interaction can be quantitatively determined by changes in fluorescent signal strength.
Spatial resolution
BiFC allows measurement of spatial and temporal changes in protein complexes, even in response to activating and inhibiting drugs and subcellularly, providing the highest
spatial resolution
In physics and geosciences, the term spatial resolution refers to distance between independent measurements, or the physical dimension that represents a pixel of the image. While in some instruments, like cameras and telescopes, spatial resolutio ...
of ''in vivo'' protein–protein interaction assays.
[Michnick, S. W., Ear, P. H., Manderson, E. N., Remy, I. & Stefan, E. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat. Rev. Drug Discov. 6, 569–582 (2007).][MacDonald, M. L. et al. Identifying off-target effects and hidden phenotypes of drugs in human cells. Nat. Chem. Biol. 2, 329–337 (2006).]
No specialised equipment
BiFC does not require specialised equipment, as visualisation is possible with an inverted fluorescence microscope that can detect fluorescence in cells.
In addition, analysis does not require complex data processing or correction for other sources of fluorescence.
No structural information needed
BiFC can be performed without structural information about the interaction partners, so long as the fluorescent reporter protein fragments can associate within the complex, as multiple combinations of fusion proteins can be screened. This is due to the assumption that, since the protein functions are recapitulated in the ''in vivo'' context, the complex structure will resemble that of the intact proteins seen physiologically.
Multiple applications
The BiFC technology has been refined and expanded to include the abilities to
simultaneously visualise multiple protein complexes in the same cell,
RNA/protein interactions, to quickly
detect changes in gene transduction pathways, demonstrate hidden phenotypes of drugs, where the predicted treatment outcome (i.e. cell death, differentiation, morphological change) is not seen ''in vivo'',
study complex formation in different cellular compartments, and
to map protein interaction surfaces[Rackham, O. & Brown, C. M. Visualization of RNA-protein interactions in living cells: FMRP and IMP1 interact on mRNAs. EMBO J. 23, 3346–3355 (2004).][Remy, I., Wilson, I. A. & Michnick, S. W. Erythropoietin receptor activation by a ligand-induced conformation change. Science 283, 990–993 (1999).][Remy, I., Montmarquette, A. & Michnick, S. W. PKB/Akt modulates TGF-beta signalling through a direct interaction with Smad3. Nat. Cell Biol. 6, 358–365 (2004)]
Limitations
Real-time detection
The fluorescent signal only is produced after the proteins have interacted, which is generally in the order of hours. Hence BiFC is unable to provide real-time detection of protein interactions. The delay for chemical reactions to generate fluorophore may also have an effect on the dynamics of complex
dissociation
Dissociation, in the wide sense of the word, is an act of disuniting or separating a complex object into parts. Dissociation may also refer to:
* Dissociation (chemistry), general process in which molecules or ionic compounds (complexes, or salts) ...
and partner exchange.
[Magliery, T. J. et al. Detecting protein–protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J. Am. Chem. Soc. 127, 146–157 (2005).]
Irreversible BiFC formation
BiFC complex formation is only reversible during the initial step of fluorescent reporter protein re-assembly, typically in the order of milliseconds. Once the fluorochrome has been reconstituted, it is essentially irreversible ''in vitro''. This prevents proteins from interacting with others and may disrupt the association/disassociation of protein complexes in
dynamic equilibrium
In chemistry, a dynamic equilibrium exists once a reversible reaction occurs. Substances transition between the reactants and products at equal rates, meaning there is no net change. Reactants and products are formed at such a rate that the conc ...
.
Independent fluorescent protein fragment associations
Fluorescent protein fragments have a limited ability to associate independent of the proteins to which they are fused. Although protein-independent association will vary depending on identities of the fusion proteins and their expression levels, one must provide the necessary and numerous controls to distinguish between true and false-positive protein interactions. Generally, this limitation is mitigated by ensuring that the fusion proteins of interest are expressed at endogenous concentrations.
Altering protein structure and steric hindrance
Fluorescent fragment linkage may alter the folding or structure of the protein of interest, leading to the elimination of an interacting protein's surface binding site. In addition, the arrangement of the fluorescent fragments may prevent fluorophore reconstitution through
steric hindrance
Steric effects arise from the spatial arrangement of atoms. When atoms come close together there is a rise in the energy of the molecule. Steric effects are nonbonding interactions that influence the shape ( conformation) and reactivity of ions ...
, although steric hindrance can be reduced or eliminated by using a linker sequence that allows sufficient flexibility for the fluorescent fragments to associate. Therefore, absence of fluorescence complementation may be a false negative and does not necessarily prove that the interaction in question does not occur.
Obligate anaerobes
Due to the requirement of molecular oxygen for fluorophore formation, BiFC cannot be used in
obligate anaerobe
Obligate anaerobes are microorganisms killed by normal atmospheric concentrations of oxygen (20.95% O2). Oxygen tolerance varies between species, with some species capable of surviving in up to 8% oxygen, while others lose viability in environm ...
s, which cannot survive in the presence of oxygen. This limits the use of BiFC to
aerobic organism
Aerobic means "requiring air," in which "air" usually means oxygen.
Aerobic may also refer to
* Aerobic exercise, prolonged exercise of moderate intensity
* Aerobics, a form of aerobic exercise
* Aerobic respiration, the aerobic process of cell ...
s.
Autofluorescence
Autofluorescence is usually not a problem since the BiFC signal will be a lot higher than the background. However certain organisms, especially
apicomplexa
The Apicomplexa (also called Apicomplexia) are a large phylum of parasitic alveolates. Most of them possess a unique form of organelle that comprises a type of non-photosynthetic plastid called an apicoplast, and an apical complex structure. T ...
, have a higher autofluorescence that make it harder to apply BiFC in them. Certain fungi, such as ''
Candida albicans
''Candida albicans'' is an opportunistic pathogenic yeast that is a common member of the human gut flora. It can also survive outside the human body. It is detected in the gastrointestinal tract and mouth in 40–60% of healthy adults. It is us ...
'', also have a high autofluorescent background, but BiFC can often still be performed when the proper controls and strains are used.
Use of fusion proteins
Because endogenous wild-type proteins cannot be visualised ''in vivo'', fusion proteins must be created and their plasmids transfected into the cells studied. These fusion proteins may not recapitulate the functions, localisation, and interactions common to their wild-type counterparts, providing an inaccurate picture of the proteins in question. This problem can be alleviated by using structural information and the location of interaction sites to rationally identify fusion sites on the proteins of interest, using appropriate controls, and comparing the expression levels and functions of the fusion and wild-type proteins through Western Blots and functional assays.
Temperature dependence
Although low temperatures favour the reconstitution of fluorescence when fragments are within proximity, this may affect the behaviour of the target proteins leading to inaccurate conclusions regarding the nature of protein interactions and their interacting partners.
[Fan, J. Y. et al. Split mCherry as a new red bimolecular fluorescence complementation system for visualizing protein–protein interactions in living cells. Biochem. Biophys. Res. Commun. 367, 47–53 (2008).]
Exact interaction relationship unknown
Because fluorophore reconstitution can occur at a distance of 7 nm or more, fluorescence complementation may indicate either a direct or indirect (i.e. within the same complex) interaction between the fluorescent fragments' fused proteins.
Application
In addition to the validation of protein–protein interactions described above, BiFC has been expanded and adapted to other applications:
Assembly of bacterial ribosomes
The BiFC system has been applied to record ribosome biogenesis events in ''E.coli''.
The process of ribosomes assembly involves nucleation of ribosomal proteins in proper order and orientation. Perturbations in assembly can lead to structural defects in ribosomal subunits which as a result cannot join in the correct orientation to form fully functional ribosomes. Thus, the events of subunit joining signaled by the appearance of BiFC is an easy way to monitor ribosome biogenesis in contrast to laborious polysome profiling methods.
Multicolour fluorescence
The fluorescent protein fragments used in BiFC have been expanded to include the colours blue, cyan, green, yellow, red, cherry, and
Venus
Venus is the second planet from the Sun. It is sometimes called Earth's "sister" or "twin" planet as it is almost as large and has a similar composition. As an interior planet to Earth, Venus (like Mercury) appears in Earth's sky never fa ...
.
[Jach,G.; Pesch,M.; Richter,K.; Frings,S.; Uhrig,J.F. An improved mRFP1 adds red to bimolecular fluorescence complementation. Nat. Methods. 3, 597–600 (2006)][Shyu,Y.J., Liu,H., Deng,X., Hu,C.D. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. BioTechniques. 40, 61–66 (2006).] This range in colours has made the development of multicolour fluorescence complementation analysis possible.
This technique allows multiple protein complexes to be visualised simultaneously in the same cell. In addition, proteins typically have a large number of alternative interaction partners. Therefore, by fusing fragments of different fluorescent proteins to candidate proteins, one can study competition between alternative interaction partners for complex formation through the complementation of different fluorescent colour fragments.
RNA-binding protein interactions
BiFC has been expanded to include the study of RNA-binding protein interactions in a method Rackham and Brown described as trimolecular fluorescence complementation (TriFC).
In this method, a fragment of the Venus fluorescent protein is fused to the
mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein.
mRNA is ...
of interest, and the complementary Venus portion fused to the
RNA-binding protein
RNA-binding proteins (often abbreviated as RBPs) are proteins that bind to the double or single stranded RNA in cells and participate in forming ribonucleoprotein complexes.
RBPs contain various structural motifs, such as RNA recognition motif ( ...
of interest. Similar to BiFC, if the mRNA and protein interact, the Venus protein will be reconstituted and fluoresce. Also known as the RNA bridge method, as the fluorophore and other interacting proteins form a bridge between the protein and the RNA of interest, this allows a simple detection and localisation of RNA-protein interactions within a living cell and provides a simple method to detect direct or indirect RNA-protein association (i.e. within a complex) that can be verified through in vitro analysis of purified compounds or
RNAi
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by o ...
knockdown of the bridging molecule(s).
Pathway organisation and signal transduction cascades
BiFC can be used to link genes to one other and their function through measurement of interactions among the proteins that the genes encode.
This application is ideal for novel genes in which little is known about their
up- and downstream effectors, as novel pathway linkages can be made. In addition, the effects of drugs,
hormone
A hormone (from the Greek participle , "setting in motion") is a class of signaling molecules in multicellular organisms that are sent to distant organs by complex biological processes to regulate physiology and behavior. Hormones are required ...
s, or
deletion or knockdown of the gene of interest, and the subsequent effects on both the strength of the protein–protein interactions and the location of the interaction can be observed within seconds.
Complex formation in different cellular compartments
BiFC has been used to study
nuclear translocation, via complex localisation, as well as interactions involving
integral membrane protein
An integral, or intrinsic, membrane protein (IMP) is a type of membrane protein that is permanently attached to the biological membrane. All ''transmembrane proteins'' are IMPs, but not all IMPs are transmembrane proteins. IMPs comprise a signi ...
s.
[de Virgilio, M., Kiosses, W. B. & Shattil, S. J. Proximal, selective, and dynamic interactions between integrin alphaIIbbeta3 and protein tyrosine kinases in living cells. J. Cell Biol. 165, 305–311 (2004).][Tong, E. H. et al. Regulation of nucleocytoplasmic trafficking of transcription factor OREBP/TonEBP/NFAT5. J. Biol. Chem. 281, 23870-23879 (2006).][Lopez-Gimenez, J. F., Canals, M., Pediani, J. D. & Milligan, G. The alpha1b-adrenoceptor exists as a higher-order oligomer: effective oligomerization is required for receptor maturation, surface delivery, and function. Mol. Pharmacol. 71, 1015–1029 (2007).][Nakahara, S., Hogan, V., Inohara, H. & Raz, A. Importin-mediated nuclear translocation of galectin-3. J. Biol. Chem. 281, 39649-39659 (2006).][Liu, H. et al. Mutual regulation of c-Jun and ATF2 by transcriptional activation and subcellular localization. EMBO J. 25, 1058–1069 (2006).][Gwozdz, T. et al. EcR and Usp, components of the ecdysteroid nuclear receptor complex, exhibit differential distribution of molecular determinants directing subcellular trafficking. Cell. Signal. 19, 490–503 (2007).][Fan, M., Ahmed, K. M., Coleman, M. C., Spitz, D. R. & Li, J. J. Nuclear factor-kappaB and manganese superoxide dismutase mediate adaptive radioresistance in low-dose irradiated mouse skin epithelial cells. Cancer Res. 67, 3220–3228 (2007).] Thus, BiFC is an important tool in understanding transcription factor localisation in subcellular compartments.
Quantifying protein–protein interaction surfaces
BiFC has been coupled with
flow cytometry
Flow cytometry (FC) is a technique used to detect and measure physical and chemical characteristics of a population of cells or particles.
In this process, a sample containing cells or particles is suspended in a fluid and injected into the flo ...
(BiFC-FC). This allows protein–protein interaction surfaces to be mapped through the introduction of
site-directed or random mutations that affect complex formation.
Comparisons to other technologies
Most techniques used to study protein–protein interactions rely on ''in vitro'' methods. Unfortunately, studying proteins in an artificial system, outside of their cellular environment, poses a number of difficulties. For example, this may require the removal of proteins from their normal cellular environment. The processing required to isolate the protein may affect its interactions with other proteins. In addition, isolating the protein from the
intracellular signaling
In biology, cell signaling (cell signalling in British English) or cell communication is the ability of a cell to receive, process, and transmit signals with its environment and with itself. Cell signaling is a fundamental property of all cellula ...
and mechanisms that occur in the normal cell may provide a misleading picture of intracellular and physiological occurrences.
Furthermore, proteins studied in vitro may be studied at concentrations vastly different from their normal abundance levels, may not necessarily be transported efficiently into the cells, or may not be selective enough to function in the host genome.
[Wu, P., Daniel-Issakani, S., LaMarco, K. & Strulovici, B. An automated high throughput filtration assay: application to polymerase inhibitor identification. Anal. Biochem. 245, 226–230 (1997).][Stoevesandt, O. & Brock, R. One-step analysis of protein complexes in microliters of cell lysate using indirect immunolabeling & fluorescence cross-correlation spectroscopy. Nat. Protoc. 1, 223–229 (2006).][Bergendahl, V., Heyduk, T. & Burgess, R. R. Luminescence resonance energy transfer-based high-throughput screening assay for inhibitors of essential protein–protein interactions in bacterial RNA polymerase. Appl. Environ. Microbiol. 69, 1492–1498 (2003).][Yang, P. et al. Multiplexed detection of protein–peptide interaction and inhibition using capillary electrophoresis. Anal. Chem. 79, 1690–1695 (2007).] Finally, by studying proteins ''in vitro'', one is unable to determine the influence of specific protein–protein interactions in the cell on the functional or physiological consequences.
Other ''in vivo'' assays most commonly used to study protein–protein interactions include
fluorescence resonance energy transfer
Fluorescence is the emission of light by a substance that has absorbed light or other electromagnetic radiation. It is a form of luminescence. In most cases, the emitted light has a longer wavelength, and therefore a lower photon energy, tha ...
(
FRET
A fret is any of the thin strips of material, usually metal wire, inserted laterally at specific positions along the neck or fretboard of a stringed instrument. Frets usually extend across the full width of the neck. On some historical instrume ...
) and
yeast two-hybrid
Two-hybrid screening (originally known as yeast two-hybrid system or Y2H) is a molecular biology technique used to discover protein–protein interactions (PPIs) and protein–DNA interactions by testing for physical interactions (such as bindi ...
(
Y2H) assay. Each of these assays has their advantages and disadvantages in comparison to BiFC:
Fluorescence resonance energy transfer (FRET)
Fluorescence resonance energy transfer
Fluorescence is the emission of light by a substance that has absorbed light or other electromagnetic radiation. It is a form of luminescence. In most cases, the emitted light has a longer wavelength, and therefore a lower photon energy, tha ...
(
FRET
A fret is any of the thin strips of material, usually metal wire, inserted laterally at specific positions along the neck or fretboard of a stringed instrument. Frets usually extend across the full width of the neck. On some historical instrume ...
), also known as
förster resonance energy transfer
Förster resonance energy transfer (FRET), fluorescence resonance energy transfer, resonance energy transfer (RET) or electronic energy transfer (EET) is a mechanism describing energy transfer between two light-sensitive molecules ( chromophores). ...
,
resonance energy transfer
Resonance describes the phenomenon of increased amplitude that occurs when the frequency of an applied Periodic function, periodic force (or a Fourier analysis, Fourier component of it) is equal or close to a natural frequency of the system ...
(
RET) or
electronic energy transfer (
EET
"Eet" is a song from Regina Spektor's fifth studio album, ''Far (album), Far''. It was released as the album's second official single in October 2009. In Europe it was released as a digital download on November 27, 2009.
Music video
A Viral vide ...
), is based on the transfer of energy from an excited (
donor
A donor in general is a person, organization or government which donates something voluntarily. The term is usually used to represent a form of pure altruism, but is sometimes used when the payment for a service is recognized by all parties as rep ...
)
chromophore
A chromophore is the part of a molecule responsible for its color.
The color that is seen by our eyes is the one not absorbed by the reflecting object within a certain wavelength spectrum of visible light. The chromophore is a region in the molec ...
or
fluorophore
A fluorophore (or fluorochrome, similarly to a chromophore) is a fluorescent chemical compound that can re-emit light upon light excitation. Fluorophores typically contain several combined aromatic groups, or planar or cyclic molecules with se ...
(if the chromophores are fluorescent) to a nearby
acceptor
Acceptor may refer to:
* Acceptor (accounting), the addressee of a bill of exchange
* In the Indian Contract Act of 1872, the acceptor is the person to whom a proposal is made, and who has communicated his or her acceptance of the said proposal
...
. In this method, fluorophores are chemically linked or genetically fused to two proteins hypothesised to interact. If the proteins interact, this will bring the fluorophores into close spatial proximity. If the fluorophores are oriented in a manner that exposes the fluorophores to one another, usually ensured when designing and constructing the fluorophore-protein linkage/fusion, then the
energy transfer
In physics, energy (from Ancient Greek: ἐνέργεια, ''enérgeia'', “activity”) is the quantitative property that is transferred to a body or to a physical system, recognizable in the performance of work and in the form of heat ...
from the excited donor fluorophore will result in a change in the fluorescent intensities or lifetimes of the fluorophores.
Yeast two-hybrid (Y2H)
The
yeast two-hybrid
Two-hybrid screening (originally known as yeast two-hybrid system or Y2H) is a molecular biology technique used to discover protein–protein interactions (PPIs) and protein–DNA interactions by testing for physical interactions (such as bindi ...
(
Y2H) is a genetic screening technique that can be used to detect physical (binding) protein–protein or
protein–DNA interactions. It is usually applied in the model yeast organism ''
Saccharomyces cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungus microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have been o ...
''. It tests a 'bait' protein of (un)known function that is fused to, for example, the binding domain of the transcription factor
GAL4
The Gal4 transcription factor is a positive regulator of gene expression of galactose-induced genes. This protein represents a large fungal family of transcription factors, Gal4 family, which includes over 50 members in the yeast ''Saccharomyces ...
against potential interacting proteins or a cDNA library that express , for example, the GAL4 activation domain (the 'prey').
[Fields, S. & Song, O. A novel genetic system to detect protein–protein interactions. Nature 340, 245–246 (1989).]
Technology comparisons
References
{{reflist
External links
Kerppola Lab Online – BiFC Dr. C-D Hu Presentation – Bimolecular Fluorescence Complementation (BiFC): Principles & Applications
Protein methods
Protein–protein interaction assays
 Bimolecular fluorescence complementation (also known as BiFC) is a technology typically used to validate
Bimolecular fluorescence complementation (also known as BiFC) is a technology typically used to validate 