|
Consensus Sequence
In molecular biology and bioinformatics, the consensus sequence (or canonical sequence) is the calculated order of most frequent residues, either nucleotide or amino acid, found at each position in a sequence alignment. It serves as a simplified representation of the population. It represents the results of multiple sequence alignments in which related sequences are compared to each other and similar sequence motifs are calculated. Such information is important when considering sequence-dependent enzymes such as RNA polymerase.Pierce, Benjamin A. 2002. Genetics : A Conceptual Approach. 1st ed. New York: W.H. Freeman and Co. Biological significance A protein binding site, represented by a consensus sequence, may be a short sequence of nucleotides which is found several times in the genome and is thought to play the same role in its different locations. For example, many transcription factors recognize particular patterns in the promoters of the genes they regulate. In the same way, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Biology
Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions. The study of chemical and physical structure of biological macromolecules is known as molecular biology. Molecular biology was first described as an approach focused on the underpinnings of biological phenomena - uncovering the structures of biological molecules as well as their interactions, and how these interactions explain observations of classical biology. In 1945 the term molecular biology was used by physicist William Astbury. In 1953 Francis Crick, James Watson, Rosalind Franklin, and colleagues, working at Medical Research Council unit, Cavendish laboratory, Cambridge (now the MRC Laboratory of Molecular Biology), made a double helix model of DNA which changed the entire research scenario. They proposed the DNA structure based on previous research done by Ro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Binding Site
DNA binding sites are a type of binding site found in DNA where other molecules may bind. DNA binding sites are distinct from other binding sites in that (1) they are part of a DNA sequence (e.g. a genome) and (2) they are bound by DNA-binding proteins. DNA binding sites are often associated with specialized proteins known as transcription factors, and are thus linked to transcriptional regulation. The sum of DNA binding sites of a specific transcription factor is referred to as its cistrome. DNA binding sites also encompasses the targets of other proteins, like restriction enzymes, site-specific recombinases (see site-specific recombination) and methyltransferases. DNA binding sites can be thus defined as short DNA sequences (typically 4 to 30 base pairs long, but up to 200 bp for recombination sites) that are specifically bound by one or more DNA-binding proteins or protein complexes. It has been reported that some binding sites have potential to undergo fast evolutionary chan ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Position-specific Scoring Matrix
A position weight matrix (PWM), also known as a position-specific weight matrix (PSWM) or position-specific scoring matrix (PSSM), is a commonly used representation of motifs (patterns) in biological sequences. PWMs are often derived from a set of aligned sequences that are thought to be functionally related and have become an important part of many software tools for computational motif discovery. Background Creation Conversion of sequence to position probability matrix A PWM has one row for each symbol of the alphabet (4 rows for nucleotides in DNA sequences or 20 rows for amino acids in protein sequences) and one column for each position in the pattern. In the first step in constructing a PWM, a basic position frequency matrix (PFM) is created by counting the occurrences of each nucleotide at each position. From the PFM, a position probability matrix (PPM) can now be created by dividing that former nucleotide count at each position by the number of sequences, thereb ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
UGENE
UGENE is computer software for bioinformatics. It works on personal computer operating systems such as Windows, macOS, or Linux. It is released as free and open-source software, under a GNU General Public License (GPL) version 2. UGENE helps biologists to analyze various biological genetics data, such as sequences, annotations, multiple alignments, phylogenetic trees, NGS assemblies, and others. The data can be stored both locally (on a personal computer) and on a shared storage (e.g., a lab database). UGENE integrates dozens of well-known biological tools, algorithms, and original tools in the context of genomics, evolutionary biology, virology, and other branches of life science. UGENE provides a graphical user interface (GUI) for the pre-built tools so biologists with no computer programming skills can access those tools more easily. Using UGENE Workflow Designer, it is possible to streamline a multi-step analysis. The workflow consists of blocks such as data readers, blocks ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Logo
In bioinformatics, a sequence logo is a graphical representation of the sequence conservation of nucleotides (in a strand of DNA/RNA) or amino acids (in protein sequences). A sequence logo is created from a collection of aligned sequences and depicts the consensus sequence and diversity of the sequences. Sequence logos are frequently used to depict sequence characteristics such as protein-binding sites in DNA or functional units in proteins. Overview A sequence logo consists of a stack of letters at each position. The relative sizes of the letters indicate their frequency in the sequences. The total height of the letters depicts the information content of the position, in bits. Logo creation To create sequence logos, related DNA, RNA or protein sequences, or DNA sequences that have common conserved binding sites, are aligned so that the most conserved parts create good alignments. A sequence logo can then be created from the conserved multiple sequence alignment. The sequence ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
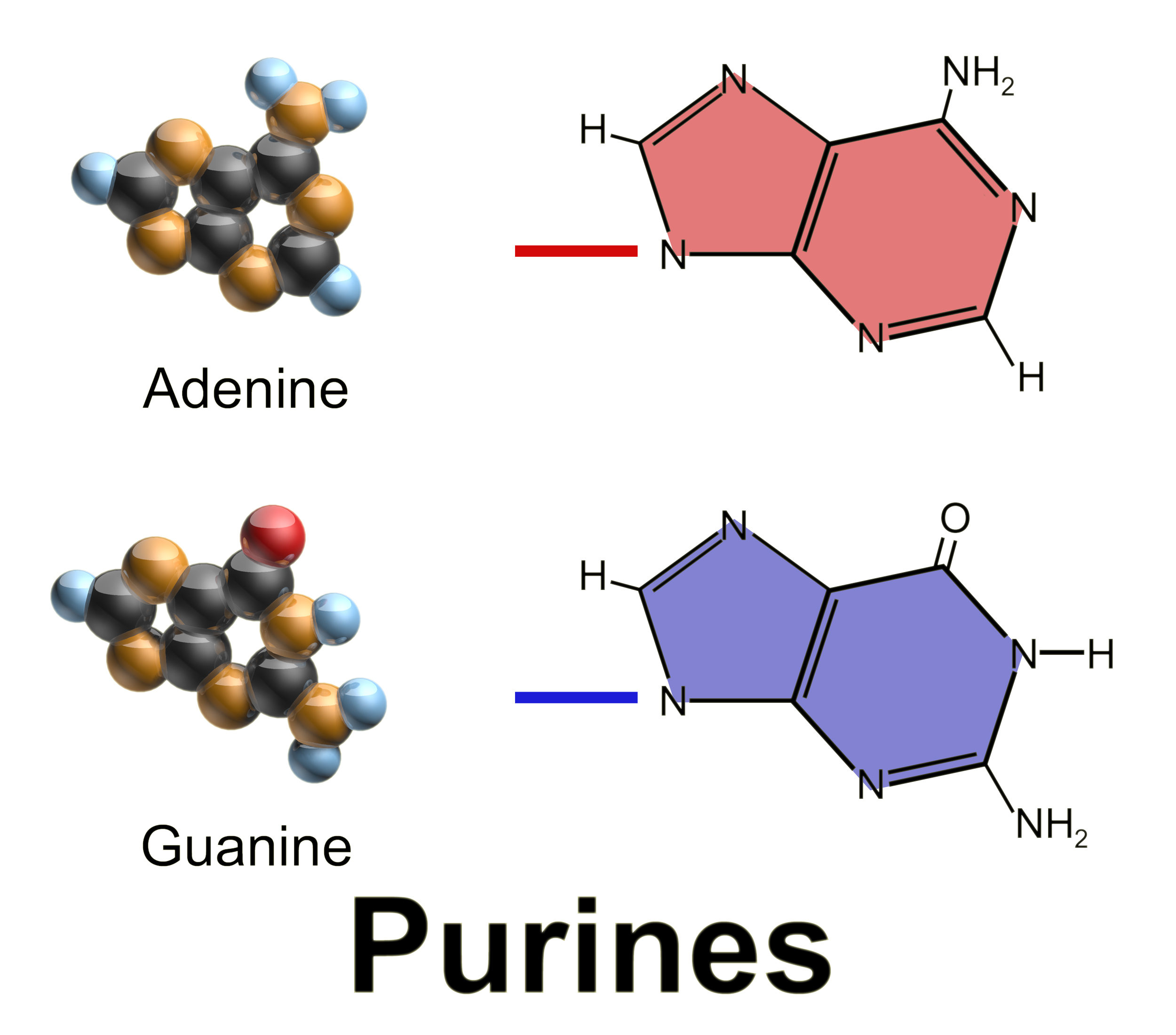
Purine
Purine is a heterocyclic compound, heterocyclic aromatic organic compound that consists of two rings (pyrimidine and imidazole) fused together. It is water-soluble. Purine also gives its name to the wider class of molecules, purines, which include substituted purines and their tautomers. They are the most widely occurring nitrogen-containing heterocycles in nature. Dietary sources Purines are found in high concentration in meat and meat products, especially internal organs such as liver and kidney. In general, plant-based diets are low in purines. High-purine plants and algae include some legumes (lentils and Black-eyed pea, black eye peas) and Spirulina (dietary supplement), spirulina. Examples of high-purine sources include: sweetbreads, Anchovies as food, anchovies, Sardines as food, sardines, liver, beef kidneys, Brain as food, brains, meat extracts (e.g., Oxo (food), Oxo, Bovril), herring, mackerel, scallops, game meats, yeast (beer, yeast extract, nutritional yeast) and g ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Pyrimidine
Pyrimidine (; ) is an aromatic, heterocyclic, organic compound similar to pyridine (). One of the three diazines (six-membered heterocyclics with two nitrogen atoms in the ring), it has nitrogen atoms at positions 1 and 3 in the ring. The other diazines are pyrazine (nitrogen atoms at the 1 and 4 positions) and pyridazine (nitrogen atoms at the 1 and 2 positions). In nucleic acids, three types of nucleobases are pyrimidine derivatives: cytosine (C), thymine (T), and uracil (U). Occurrence and history The pyrimidine ring system has wide occurrence in nature as substituted and ring fused compounds and derivatives, including the nucleotides cytosine, thymine and uracil, thiamine (vitamin B1) and alloxan. It is also found in many synthetic compounds such as barbiturates and the HIV drug, zidovudine. Although pyrimidine derivatives such as alloxan were known in the early 19th century, a laboratory synthesis of a pyrimidine was not carried out until 1879, when Grimaux reported the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Motif
In biology, a sequence motif is a nucleotide or amino-acid sequence pattern that is widespread and usually assumed to be related to biological function of the macromolecule. For example, an ''N''-glycosylation site motif can be defined as ''Asn, followed by anything but Pro, followed by either Ser or Thr, followed by anything but Pro residue''. Overview When a sequence motif appears in the exon of a gene, it may encode the "structural motif" of a protein; that is a stereotypical element of the overall structure of the protein. Nevertheless, motifs need not be associated with a distinctive secondary structure. " Noncoding" sequences are not translated into proteins, and nucleic acids with such motifs need not deviate from the typical shape (e.g. the "B-form" DNA double helix). Outside of gene exons, there exist regulatory sequence motifs and motifs within the " junk", such as satellite DNA. Some of these are believed to affect the shape of nucleic acids (see for example RN ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Evolution
Evolution is change in the heritable characteristics of biological populations over successive generations. These characteristics are the expressions of genes, which are passed on from parent to offspring during reproduction. Variation tends to exist within any given population as a result of genetic mutation and recombination. Evolution occurs when evolutionary processes such as natural selection (including sexual selection) and genetic drift act on this variation, resulting in certain characteristics becoming more common or more rare within a population. The evolutionary pressures that determine whether a characteristic is common or rare within a population constantly change, resulting in a change in heritable characteristics arising over successive generations. It is this process of evolution that has given rise to biodiversity at every level of biological organisation, including the levels of species, individual organisms, and molecules. The theory of evolution by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Signal Peptide
A signal peptide (sometimes referred to as signal sequence, targeting signal, localization signal, localization sequence, transit peptide, leader sequence or leader peptide) is a short peptide (usually 16-30 amino acids long) present at the N-terminus (or occasionally nonclassically at the C-terminus or internally) of most newly synthesized proteins that are destined toward the secretory pathway. These proteins include those that reside either inside certain organelles (the endoplasmic reticulum, Golgi or endosomes), secreted from the cell, or inserted into most cellular membranes. Although most type I membrane-bound proteins have signal peptides, the majority of type II and multi-spanning membrane-bound proteins are targeted to the secretory pathway by their first transmembrane domain, which biochemically resembles a signal sequence except that it is not cleaved. They are a kind of target peptide. Function (translocation) Signal peptides function to prompt a cell to translo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Regulatory Sequence
A regulatory sequence is a segment of a nucleic acid molecule which is capable of increasing or decreasing the expression of specific genes within an organism. Regulation of gene expression is an essential feature of all living organisms and viruses. Description In DNA, regulation of gene expression normally happens at the level of RNA biosynthesis (transcription). It is accomplished through the sequence-specific binding of proteins (transcription factors) that activate or inhibit transcription. Transcription factors may act as activators, repressors, or both. Repressors often act by preventing RNA polymerase from forming a productive complex with the transcriptional initiation region ( promoter), while activators facilitate formation of a productive complex. Furthermore, DNA motifs have been shown to be predictive of epigenomic modifications, suggesting that transcription factors play a role in regulating the epigenome. In RNA, regulation may occur at the level of protein ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |