histone modification on:
[Wikipedia]
[Google]
[Amazon]
In
 Five major families of histone proteins exist: H1/H5, H2A, H2B, H3, and H4. Histones H2A, H2B, H3 and H4 are known as the core or nucleosomal histones, while histones H1/H5 are known as the linker histones.
The core histones all exist as dimers, which are similar in that they all possess the histone fold domain: three alpha helices linked by two loops. It is this helical structure that allows for interaction between distinct dimers, particularly in a head-tail fashion (also called the handshake motif). The resulting four distinct dimers then come together to form one octameric
Five major families of histone proteins exist: H1/H5, H2A, H2B, H3, and H4. Histones H2A, H2B, H3 and H4 are known as the core or nucleosomal histones, while histones H1/H5 are known as the linker histones.
The core histones all exist as dimers, which are similar in that they all possess the histone fold domain: three alpha helices linked by two loops. It is this helical structure that allows for interaction between distinct dimers, particularly in a head-tail fashion (also called the handshake motif). The resulting four distinct dimers then come together to form one octameric
"HistoneDB 2.0 - Variants"
database. Several pseudogenes have also been discovered and identified in very close sequences of their respective functional ortholog genes. The following is a list of human histone proteins, genes and pseudogenes:
 The
The

 Histones undergo
Histones undergo
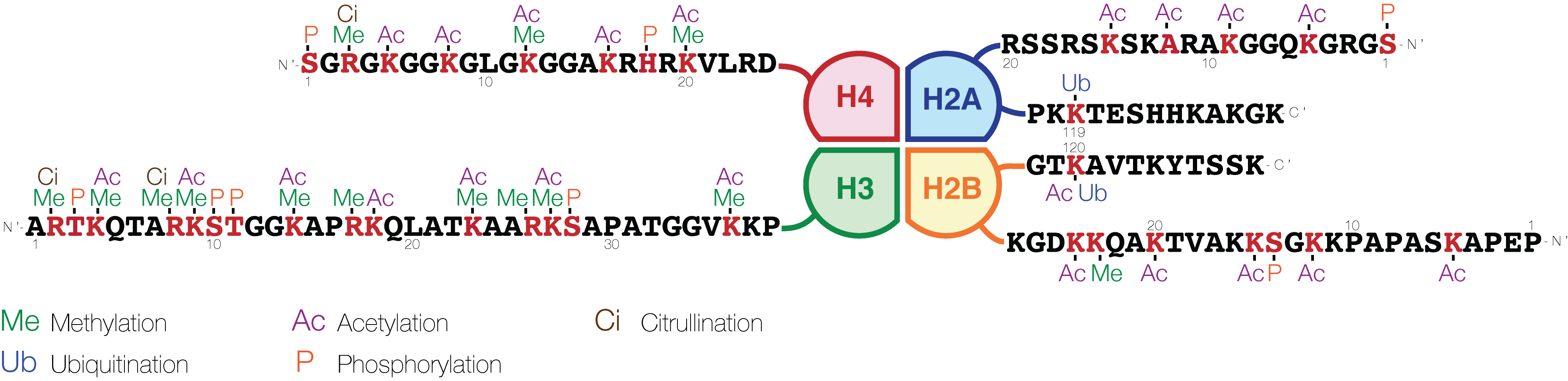 A huge catalogue of histone modifications have been described, but a functional understanding of most is still lacking. Collectively, it is thought that histone modifications may underlie a
A huge catalogue of histone modifications have been described, but a functional understanding of most is still lacking. Collectively, it is thought that histone modifications may underlie a
HistoneDB 2.0 - Database of histones and variants
at
Chromatin, Histones & Cathepsin
PMAP The Proteolysis Map-animation {{Authority control Epigenetics Proteins DNA-binding proteins
biology
Biology is the scientific study of life and living organisms. It is a broad natural science that encompasses a wide range of fields and unifying principles that explain the structure, function, growth, History of life, origin, evolution, and ...
, histones are highly basic
Basic or BASIC may refer to:
Science and technology
* BASIC, a computer programming language
* Basic (chemistry), having the properties of a base
* Basic access authentication, in HTTP
Entertainment
* Basic (film), ''Basic'' (film), a 2003 film
...
protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
s abundant in lysine
Lysine (symbol Lys or K) is an α-amino acid that is a precursor to many proteins. Lysine contains an α-amino group (which is in the protonated form when the lysine is dissolved in water at physiological pH), an α-carboxylic acid group ( ...
and arginine
Arginine is the amino acid with the formula (H2N)(HN)CN(H)(CH2)3CH(NH2)CO2H. The molecule features a guanidinium, guanidino group appended to a standard amino acid framework. At physiological pH, the carboxylic acid is deprotonated (−CO2−) a ...
residues that are found in eukaryotic
The eukaryotes ( ) constitute the Domain (biology), domain of Eukaryota or Eukarya, organisms whose Cell (biology), cells have a membrane-bound cell nucleus, nucleus. All animals, plants, Fungus, fungi, seaweeds, and many unicellular organisms ...
cell nuclei
The cell nucleus (; : nuclei) is a membrane-bound organelle found in eukaryotic cells. Eukaryotic cells usually have a single nucleus, but a few cell types, such as mammalian red blood cells, have no nuclei, and a few others including osteoc ...
and in most Archaeal phyla
Phyla, the plural of ''phylum'', may refer to:
* Phylum, a biological taxon between Kingdom and Class
* by analogy, in linguistics, a large division of possibly related languages, or a major language family which is not subordinate to another
Phy ...
. They act as spools around which DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
winds to create structural units called nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone, histone proteins and resembles thread wrapped around a bobbin, spool. The nucleosome ...
s. Nucleosomes in turn are wrapped into 30-nanometer
330px, Different lengths as in respect to the Molecule">molecular scale.
The nanometre (international spelling as used by the International Bureau of Weights and Measures; SI symbol: nm), or nanometer (American spelling
Despite the va ...
fibers that form tightly packed chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important r ...
. Histones prevent DNA from becoming tangled and protect it from DNA damage. In addition, histones play important roles in gene regulation
Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products (protein or RNA). Sophisticated programs of gene expression are wide ...
and DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all life, living organisms, acting as the most essential part of heredity, biolog ...
. Without histones, unwound DNA in chromosome
A chromosome is a package of DNA containing part or all of the genetic material of an organism. In most chromosomes, the very long thin DNA fibers are coated with nucleosome-forming packaging proteins; in eukaryotic cells, the most import ...
s would be very long. For example, each human cell has about 1.8 meters of DNA if completely stretched out; however, when wound about histones, this length is reduced to about 9 micrometers (0.09 mm) of 30 nm diameter chromatin fibers.
There are five families of histones, which are designated H1/H5 (linker histones), H2, H3, and H4 (core histones). The nucleosome core is formed of two H2A-H2B dimers and a H3-H4 tetramer
A tetramer () (''tetra-'', "four" + '' -mer'', "parts") is an oligomer formed from four monomers or subunits. The associated property is called ''tetramery''. An example from inorganic chemistry is titanium methoxide with the empirical formula ...
. The tight wrapping of DNA around histones, is to a large degree, a result of electrostatic
Electrostatics is a branch of physics that studies slow-moving or stationary electric charges.
Since classical times, it has been known that some materials, such as amber, attract lightweight particles after rubbing. The Greek word (), mean ...
attraction between the positively charged histones and negatively charged phosphate backbone of DNA.
Histones may be chemically modified through the action of enzymes to regulate gene transcription. The most common modifications are the methylation
Methylation, in the chemistry, chemical sciences, is the addition of a methyl group on a substrate (chemistry), substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replac ...
of arginine or lysine residues or the acetylation
:
In chemistry, acetylation is an organic esterification reaction with acetic acid. It introduces an acetyl group into a chemical compound. Such compounds are termed ''acetate esters'' or simply ''acetates''. Deacetylation is the opposite react ...
of lysine. Methylation can affect how other proteins such as transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription (genetics), transcription of genetics, genetic information from DNA to messenger RNA, by binding t ...
s interact with the nucleosomes. Lysine acetylation eliminates a positive charge on lysine thereby weakening the electrostatic attraction between histone and DNA, resulting in partial unwinding of the DNA, making it more accessible for gene expression.
Classes and variants
 Five major families of histone proteins exist: H1/H5, H2A, H2B, H3, and H4. Histones H2A, H2B, H3 and H4 are known as the core or nucleosomal histones, while histones H1/H5 are known as the linker histones.
The core histones all exist as dimers, which are similar in that they all possess the histone fold domain: three alpha helices linked by two loops. It is this helical structure that allows for interaction between distinct dimers, particularly in a head-tail fashion (also called the handshake motif). The resulting four distinct dimers then come together to form one octameric
Five major families of histone proteins exist: H1/H5, H2A, H2B, H3, and H4. Histones H2A, H2B, H3 and H4 are known as the core or nucleosomal histones, while histones H1/H5 are known as the linker histones.
The core histones all exist as dimers, which are similar in that they all possess the histone fold domain: three alpha helices linked by two loops. It is this helical structure that allows for interaction between distinct dimers, particularly in a head-tail fashion (also called the handshake motif). The resulting four distinct dimers then come together to form one octameric nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone, histone proteins and resembles thread wrapped around a bobbin, spool. The nucleosome ...
core, approximately 63 Angstroms in diameter (a solenoid (DNA)-like particle). Around 146 base pairs
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
(bp) of DNA wrap around this core particle 1.65 times in a left-handed super-helical turn to give a particle of around 100 Angstroms across. The linker histone H1 binds the nucleosome at the entry and exit sites of the DNA, thus locking the DNA into place and allowing the formation of higher order structure. The most basic such formation is the 10 nm fiber or beads on a string conformation. This involves the wrapping of DNA around nucleosomes with approximately 50 base pairs of DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
separating each pair of nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone, histone proteins and resembles thread wrapped around a bobbin, spool. The nucleosome ...
s (also referred to as linker DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
). Higher-order structures include the 30 nm fiber (forming an irregular zigzag) and 100 nm fiber, these being the structures found in normal cells. During mitosis and meiosis, the condensed chromosome
A chromosome is a package of DNA containing part or all of the genetic material of an organism. In most chromosomes, the very long thin DNA fibers are coated with nucleosome-forming packaging proteins; in eukaryotic cells, the most import ...
s are assembled through interactions between nucleosomes and other regulatory proteins.
Histones are subdivided into canonical replication-dependent histones, whose genes are expressed during the S-phase
S phase (Synthesis phase) is the phase of the cell cycle in which DNA is DNA replication, replicated, occurring between G1 phase, G1 phase and G2 phase, G2 phase. Since accurate duplication of the genome is critical to successful cell division, ...
of the cell cycle
The cell cycle, or cell-division cycle, is the sequential series of events that take place in a cell (biology), cell that causes it to divide into two daughter cells. These events include the growth of the cell, duplication of its DNA (DNA re ...
and replication-independent histone variants, expressed during the whole cell cycle. In mammals, genes encoding canonical histones are typically clustered along chromosomes in 4 different highly- conserved loci, lack intron
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e., a region inside a gene."The notion of the cistron .e., gen ...
s and use a stem loop structure at the 3' end
Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. In a single strand of DNA or RNA, the chemical convention of naming carbon atoms in the nucleotide pentose-sugar-ri ...
instead of a polyA tail
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In euka ...
. Genes encoding histone variants are usually not clustered, have introns and their mRNAs are regulated with polyA tails. Complex multicellular organisms typically have a higher number of histone variants providing a variety of different functions. Functionally, histone variants contribute to transcriptional control, epigenetic memory, and DNA repair, serving specialized functions beyond nucleosome packaging which plays distinct roles in chromatin dynamics. For example, H2A.Z is enriched at regulatory elements and promoters of actively transcribed genes, where it modulates nucleosome stability and transcription factor binding. In contrast, H3.3, a replacement variant of Histone H3
Histone H3 is one of the five main histones involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and a long N-terminal end, N-terminal tail, H3 is involved with the structure of the nucleosomes of the 'b ...
, is associated with active transcription and is preferentially deposited at enhancer elements and transcribed gene bodies. Another critical variant, CENPA
Centromere protein A, also known as CENPA, is a protein which in humans is encoded by the ''CENPA'' gene. CENPA is a histone H3 variant which is the critical factor determining the kinetochore position(s) on each chromosome in most eukaryotes i ...
, replaces H3 in centromeric nucleosomes, providing a structural foundation essential for chromosome segregation.
Variants also play essential roles in DNA repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is cons ...
. Variants such as H2A.X are phosphorylated at sites of DNA damage, marking regions for recruitment of repair proteins. This modification, commonly referred to as γH2A.X, serves as a key signal in the cellular response to double-strand breaks
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is constantly modified ...
, facilitating efficient DNA repair processes. Defects in histone variant regulation have been linked to genome instability
Genome instability (also genetic instability or genomic instability) refers to a high frequency of mutations within the genome of a cellular lineage. These mutations can include changes in nucleic acid sequences, chromosomal rearrangements or ...
, a hallmark of many cancers and age-related diseases.
Recent data are accumulating about the roles of diverse histone variants highlighting the functional links between variants and the delicate regulation of organism development. Histone variants proteins from different organisms, their classification and variant specific features can be found i"HistoneDB 2.0 - Variants"
database. Several pseudogenes have also been discovered and identified in very close sequences of their respective functional ortholog genes. The following is a list of human histone proteins, genes and pseudogenes:
Structure
nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone, histone proteins and resembles thread wrapped around a bobbin, spool. The nucleosome ...
core is formed of two H2A-H2B dimers and a H3-H4 tetramer, forming two nearly symmetrical halves by tertiary structure
Protein tertiary structure is the three-dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains and the ...
( C2 symmetry; one macromolecule
A macromolecule is a "molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass." Polymers are physi ...
is the mirror image of the other). The H2A-H2B dimers and H3-H4 tetramer also show pseudodyad symmetry. The 4 'core' histones (H2A, H2B, H3 and H4) are relatively similar in structure and are highly conserved through evolution
Evolution is the change in the heritable Phenotypic trait, characteristics of biological populations over successive generations. It occurs when evolutionary processes such as natural selection and genetic drift act on genetic variation, re ...
, all featuring a ' helix turn helix turn helix' motif (DNA-binding protein motif that recognize specific DNA sequence). They also share the feature of long 'tails' on one end of the amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
structure - this being the location of post-translational modification (see below).
Archaeal histone only contains a H3-H4 like dimeric structure made out of a single type of unit. Such dimeric structures can stack into a tall superhelix ("hypernucleosome") onto which DNA coils in a manner similar to nucleosome spools. Only some archaeal histones have tails.
The distance between the spools around which eukaryotic cells wind their DNA has been determined to range from 59 to 70 Å.
In all, histones make five types of interactions with DNA:
* Salt bridges and hydrogen bonds between side chains of basic amino acids (especially lysine
Lysine (symbol Lys or K) is an α-amino acid that is a precursor to many proteins. Lysine contains an α-amino group (which is in the protonated form when the lysine is dissolved in water at physiological pH), an α-carboxylic acid group ( ...
and arginine
Arginine is the amino acid with the formula (H2N)(HN)CN(H)(CH2)3CH(NH2)CO2H. The molecule features a guanidinium, guanidino group appended to a standard amino acid framework. At physiological pH, the carboxylic acid is deprotonated (−CO2−) a ...
) and phosphate oxygens on DNA
* Helix-dipoles form alpha-helixes in H2B, H3, and H4 cause a net positive charge to accumulate at the point of interaction with negatively charged phosphate
Phosphates are the naturally occurring form of the element phosphorus.
In chemistry, a phosphate is an anion, salt, functional group or ester derived from a phosphoric acid. It most commonly means orthophosphate, a derivative of orthop ...
groups on DNA
* Hydrogen bonds
In chemistry, a hydrogen bond (H-bond) is a specific type of molecular interaction that exhibits partial covalent character and cannot be described as a purely electrostatic force. It occurs when a hydrogen (H) atom, covalently bonded to a mo ...
between the DNA backbone and the amide
In organic chemistry, an amide, also known as an organic amide or a carboxamide, is a chemical compound, compound with the general formula , where R, R', and R″ represent any group, typically organyl functional group, groups or hydrogen at ...
group on the main chain of histone proteins
* Nonpolar interactions between the histone and deoxyribose
Deoxyribose, or more precisely 2-deoxyribose, is a monosaccharide with idealized formula H−(C=O)−(CH2)−(CHOH)3−H. Its name indicates that it is a deoxy sugar, meaning that it is derived from the sugar ribose by loss of a hydroxy group. D ...
sugars on DNA
* Non-specific minor groove insertions of the H3 and H2B N-terminal tails into two minor grooves each on the DNA molecule
The highly basic nature of histones, aside from facilitating DNA-histone interactions, contributes to their water solubility.
Histones are subject to post translational modification by enzymes primarily on their N-terminal tails, but also in their globular domains. Such modifications include methylation
Methylation, in the chemistry, chemical sciences, is the addition of a methyl group on a substrate (chemistry), substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replac ...
, citrullination, acetylation
:
In chemistry, acetylation is an organic esterification reaction with acetic acid. It introduces an acetyl group into a chemical compound. Such compounds are termed ''acetate esters'' or simply ''acetates''. Deacetylation is the opposite react ...
, phosphorylation
In biochemistry, phosphorylation is described as the "transfer of a phosphate group" from a donor to an acceptor. A common phosphorylating agent (phosphate donor) is ATP and a common family of acceptor are alcohols:
:
This equation can be writ ...
, SUMOylation, ubiquitin
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 19 ...
ation, and ADP-ribosylation. This affects their function of gene regulation.
In general, gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei ...
s that are active have less bound histone, while inactive genes are highly associated with histones during interphase
Interphase is the active portion of the cell cycle that includes the G1, S, and G2 phases, where the cell grows, replicates its DNA, and prepares for mitosis, respectively. Interphase was formerly called the "resting phase," but the cell i ...
. It also appears that the structure of histones has been evolution
Evolution is the change in the heritable Phenotypic trait, characteristics of biological populations over successive generations. It occurs when evolutionary processes such as natural selection and genetic drift act on genetic variation, re ...
arily conserved, as any deleterious mutations
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mitosi ...
would be severely maladaptive. All histones have a highly positively charged N-terminus with many lysine
Lysine (symbol Lys or K) is an α-amino acid that is a precursor to many proteins. Lysine contains an α-amino group (which is in the protonated form when the lysine is dissolved in water at physiological pH), an α-carboxylic acid group ( ...
and arginine
Arginine is the amino acid with the formula (H2N)(HN)CN(H)(CH2)3CH(NH2)CO2H. The molecule features a guanidinium, guanidino group appended to a standard amino acid framework. At physiological pH, the carboxylic acid is deprotonated (−CO2−) a ...
residues.
Evolution and species distribution
Core histones are found in the nuclei ofeukaryotic
The eukaryotes ( ) constitute the Domain (biology), domain of Eukaryota or Eukarya, organisms whose Cell (biology), cells have a membrane-bound cell nucleus, nucleus. All animals, plants, Fungus, fungi, seaweeds, and many unicellular organisms ...
cells and in most Archaea
Archaea ( ) is a Domain (biology), domain of organisms. Traditionally, Archaea only included its Prokaryote, prokaryotic members, but this has since been found to be paraphyletic, as eukaryotes are known to have evolved from archaea. Even thou ...
l phyla, but not in bacteria
Bacteria (; : bacterium) are ubiquitous, mostly free-living organisms often consisting of one Cell (biology), biological cell. They constitute a large domain (biology), domain of Prokaryote, prokaryotic microorganisms. Typically a few micr ...
. The unicellular algae known as dinoflagellate
The Dinoflagellates (), also called Dinophytes, are a monophyletic group of single-celled eukaryotes constituting the phylum Dinoflagellata and are usually considered protists. Dinoflagellates are mostly marine plankton, but they are also commo ...
s were previously thought to be the only eukaryotes that completely lack histones, but later studies showed that their DNA still encodes histone genes. Unlike the core histones, homologs of the lysine-rich linker histone (H1) proteins are found in bacteria, otherwise known as nucleoprotein HC1/HC2.
It has been proposed that core histone proteins are evolutionarily related to the helical part of the extended AAA+ ATPase domain, the C-domain, and to the N-terminal substrate recognition domain of Clp/Hsp100 proteins. Despite the differences in their topology, these three folds share a homologous helix-strand-helix (HSH) motif. It's also proposed that they may have evolved from ribosomal proteins ( RPS6/ RPS15), both being short and basic proteins.
Archaeal histones may well resemble the evolutionary precursors to eukaryotic histones. Histone proteins are among the most highly conserved proteins in eukaryotes, emphasizing their important role in the biology of the nucleus. In contrast mature sperm cells largely use protamines to package their genomic DNA, most likely because this allows them to achieve an even higher packaging ratio.
There are some ''variant'' forms in some of the major classes. They share amino acid sequence homology and core structural similarity to a specific class of major histones but also have their own feature that is distinct from the major histones. These ''minor histones'' usually carry out specific functions of the chromatin metabolism. For example, histone H3-like CENPA
Centromere protein A, also known as CENPA, is a protein which in humans is encoded by the ''CENPA'' gene. CENPA is a histone H3 variant which is the critical factor determining the kinetochore position(s) on each chromosome in most eukaryotes i ...
is associated with only the centromere
The centromere links a pair of sister chromatids together during cell division. This constricted region of chromosome connects the sister chromatids, creating a short arm (p) and a long arm (q) on the chromatids. During mitosis, spindle fiber ...
region of the chromosome. Histone H2A variant H2A.Z is associated with the promoters of actively transcribed genes and also involved in the prevention of the spread of silent heterochromatin
Heterochromatin is a tightly packed form of DNA or '' condensed DNA'', which comes in multiple varieties. These varieties lie on a continuum between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a rol ...
. Furthermore, H2A.Z has roles in chromatin for genome stability. Another H2A variant H2A.X is phosphorylated at S139 in regions around double-strand breaks
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is constantly modified ...
and marks the region undergoing DNA repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is cons ...
. Histone H3.3 is associated with the body of actively transcribed genes.
Function
Compacting DNA strands
Histones act as spools around which DNA winds. This enables the compaction necessary to fit the largegenome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as ...
s of eukaryotes inside cell nuclei: the compacted molecule is 40,000 times shorter than an unpacked molecule.
Chromatin regulation
posttranslational modification
In molecular biology, post-translational modification (PTM) is the covalent process of changing proteins following protein biosynthesis. PTMs may involve enzymes or occur spontaneously. Proteins are created by ribosomes, which translate mRNA ...
s that alter their interaction with DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
and nuclear proteins. The H3 and H4 histones have long tails protruding from the nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone, histone proteins and resembles thread wrapped around a bobbin, spool. The nucleosome ...
, which can be covalent
A covalent bond is a chemical bond that involves the sharing of electrons to form electron pairs between atoms. These electron pairs are known as shared pairs or bonding pairs. The stable balance of attractive and repulsive forces between atom ...
ly modified at several places. Modifications of the tail include methylation
Methylation, in the chemistry, chemical sciences, is the addition of a methyl group on a substrate (chemistry), substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replac ...
, acetylation
:
In chemistry, acetylation is an organic esterification reaction with acetic acid. It introduces an acetyl group into a chemical compound. Such compounds are termed ''acetate esters'' or simply ''acetates''. Deacetylation is the opposite react ...
, phosphorylation
In biochemistry, phosphorylation is described as the "transfer of a phosphate group" from a donor to an acceptor. A common phosphorylating agent (phosphate donor) is ATP and a common family of acceptor are alcohols:
:
This equation can be writ ...
, ubiquitination
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 19 ...
, SUMOylation, citrullination, and ADP-ribosylation. The core of the histones H2A and H2B can also be modified. Combinations of modifications, known as ''histone marks'', are thought to constitute a code, the so-called "histone code
The histone code is a hypothesis that the transcription of genetic information encoded in DNA is in part regulated by chemical modifications (known as ''histone marks'') to histone proteins, primarily on their unstructured ends. Together with sim ...
". Histone modifications act in diverse biological processes such as gene regulation
Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products (protein or RNA). Sophisticated programs of gene expression are wide ...
, DNA repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is cons ...
, chromosome condensation (mitosis
Mitosis () is a part of the cell cycle in eukaryote, eukaryotic cells in which replicated chromosomes are separated into two new Cell nucleus, nuclei. Cell division by mitosis is an equational division which gives rise to genetically identic ...
) and spermatogenesis
Spermatogenesis is the process by which haploid spermatozoa develop from germ cells in the seminiferous tubules of the testicle. This process starts with the Mitosis, mitotic division of the stem cells located close to the basement membrane of ...
(meiosis
Meiosis () is a special type of cell division of germ cells in sexually-reproducing organisms that produces the gametes, the sperm or egg cells. It involves two rounds of division that ultimately result in four cells, each with only one c ...
).
The common nomenclature of histone modifications is:
* The name of the histone (e.g., H3)
* The single-letter amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
abbreviation (e.g., K for Lysine
Lysine (symbol Lys or K) is an α-amino acid that is a precursor to many proteins. Lysine contains an α-amino group (which is in the protonated form when the lysine is dissolved in water at physiological pH), an α-carboxylic acid group ( ...
) and the amino acid position in the protein
* The type of modification (Me: methyl
In organic chemistry, a methyl group is an alkyl derived from methane, containing one carbon atom bonded to three hydrogen atoms, having chemical formula (whereas normal methane has the formula ). In formulas, the group is often abbreviated as ...
, P: phosphate
Phosphates are the naturally occurring form of the element phosphorus.
In chemistry, a phosphate is an anion, salt, functional group or ester derived from a phosphoric acid. It most commonly means orthophosphate, a derivative of orthop ...
, Ac: acetyl
In organic chemistry, an acetyl group is a functional group denoted by the chemical formula and the structure . It is sometimes represented by the symbol Ac (not to be confused with the element actinium). In IUPAC nomenclature, an acetyl grou ...
, Ub: ubiquitin
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 19 ...
)
* The number of modifications (only Me is known to occur in more than one copy per residue. 1, 2 or 3 is mono-, di- or tri-methylation)
So H3K4me1
H3K4me1 is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the mono-methylation at the 4th lysine residue of the histone H3 protein and often associated with gene enhancers.
Nomenclature
H3K4me1 i ...
denotes the monomethylation of the 4th residue (a lysine) from the start (i.e., the N-terminal
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amin ...
) of the H3 protein.
Modification
 A huge catalogue of histone modifications have been described, but a functional understanding of most is still lacking. Collectively, it is thought that histone modifications may underlie a
A huge catalogue of histone modifications have been described, but a functional understanding of most is still lacking. Collectively, it is thought that histone modifications may underlie a histone code
The histone code is a hypothesis that the transcription of genetic information encoded in DNA is in part regulated by chemical modifications (known as ''histone marks'') to histone proteins, primarily on their unstructured ends. Together with sim ...
, whereby combinations of histone modifications have specific meanings. However, most functional data concerns individual prominent histone modifications that are biochemically amenable to detailed study.
Chemistry
Lysine methylation
The addition of one, two, or many methyl groups to lysine has little effect on the chemistry of the histone; methylation leaves the charge of the lysine intact and adds a minimal number of atoms so steric interactions are mostly unaffected. However, proteins containing Tudor, chromo or PHD domains, amongst others, can recognise lysine methylation with exquisite sensitivity and differentiate mono, di and tri-methyl lysine, to the extent that, for some lysines (e.g.: H4K20) mono, di and tri-methylation appear to have different meanings. Because of this, lysine methylation tends to be a very informative mark and dominates the known histone modification functions.Glutamine serotonylation
Recently it has been shown, that the addition of aserotonin
Serotonin (), also known as 5-hydroxytryptamine (5-HT), is a monoamine neurotransmitter with a wide range of functions in both the central nervous system (CNS) and also peripheral tissues. It is involved in mood, cognition, reward, learning, ...
group to the position 5 glutamine of H3, happens in serotonergic cells such as neurons. This is part of the differentiation of the serotonergic cells. This post-translational modification happens in conjunction with the H3K4me3 modification. The serotonylation potentiates the binding of the general transcription factor TFIID to the TATA box.
Arginine methylation
What was said above of the chemistry of lysine methylation also applies to arginine methylation, and some protein domains—e.g., Tudor domains—can be specific for methyl arginine instead of methyl lysine. Arginine is known to be mono- or di-methylated, and methylation can be symmetric or asymmetric, potentially with different meanings.Arginine citrullination
Enzymes called peptidylarginine deiminases (PADs) hydrolyze the imine group of arginines and attach a keto group, so that there is one less positive charge on the amino acid residue. This process has been involved in the activation of gene expression by making the modified histones less tightly bound to DNA and thus making the chromatin more accessible. PADs can also produce the opposite effect by removing or inhibiting mono-methylation of arginine residues on histones and thus antagonizing the positive effect arginine methylation has on transcriptional activity.Lysine acetylation
Addition of an acetyl group has a major chemical effect on lysine as it neutralises the positive charge. This reduces electrostatic attraction between the histone and the negatively charged DNA backbone, loosening the chromatin structure; highly acetylated histones form more accessible chromatin and tend to be associated with active transcription. Lysine acetylation appears to be less precise in meaning than methylation, in that histone acetyltransferases tend to act on more than one lysine; presumably this reflects the need to alter multiple lysines to have a significant effect on chromatin structure. The modification includes H3K27ac.Serine/threonine/tyrosine phosphorylation
Addition of a negatively charged phosphate group can lead to major changes in protein structure, leading to the well-characterised role ofphosphorylation
In biochemistry, phosphorylation is described as the "transfer of a phosphate group" from a donor to an acceptor. A common phosphorylating agent (phosphate donor) is ATP and a common family of acceptor are alcohols:
:
This equation can be writ ...
in controlling protein function. It is not clear what structural implications histone phosphorylation has, but histone phosphorylation has clear functions as a post-translational modification, and binding domains such as BRCT have been characterised.
Effects on transcription
Most well-studied histone modifications are involved in control of transcription.Actively transcribed genes
Two histone modifications are particularly associated with active transcription: ;''Trimethylation of H3 lysine 4 (H3K4me3)'': This trimethylation occurs at the promoter of active genes and is performed by the COMPASS complex. Despite the conservation of this complex and histone modification from yeast to mammals, it is not entirely clear what role this modification plays. However, it is an excellent mark of active promoters and the level of this histone modification at a gene's promoter is broadly correlated with transcriptional activity of the gene. The formation of this mark is tied to transcription in a rather convoluted manner: early in transcription of a gene,RNA polymerase II
RNA polymerase II (RNAP II and Pol II) is a Protein complex, multiprotein complex that Transcription (biology), transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNA pol ...
undergoes a switch from initiating' to 'elongating', marked by a change in the phosphorylation states of the RNA polymerase II C terminal domain (CTD). The same enzyme that phosphorylates
In biochemistry, phosphorylation is described as the "transfer of a phosphate group" from a donor to an acceptor. A common phosphorylating agent (phosphate donor) is ATP and a common family of acceptor are alcohols:
:
This equation can be writt ...
the CTD also phosphorylates the Rad6 complex, which in turn adds a ubiquitin mark to H2B K123 (K120 in mammals). H2BK123Ub occurs throughout transcribed regions, but this mark is required for COMPASS to trimethylate H3K4 at promoters.
;''Trimethylation of H3 lysine 36 (H3K36me3
H3K36me3 is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the tri-methylation at the 36th lysine residue of the histone H3 protein and often associated with gene, gene bodies.
There are diverse mo ...
)'': This trimethylation occurs in the body of active genes and is deposited by the methyltransferase Set2. This protein associates with elongating RNA polymerase II
RNA polymerase II (RNAP II and Pol II) is a Protein complex, multiprotein complex that Transcription (biology), transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNA pol ...
, and H3K36Me3 is indicative of actively transcribed genes. H3K36Me3 is recognised by the Rpd3 histone deacetylase complex, which removes acetyl modifications from surrounding histones, increasing chromatin compaction and repressing spurious transcription. Increased chromatin compaction prevents transcription factors from accessing DNA, and reduces the likelihood of new transcription events being initiated within the body of the gene. This process therefore helps ensure that transcription is not interrupted.
Repressed genes
Three histone modifications are particularly associated with repressed genes: ;''Trimethylation of H3 lysine 27 (H3K27me3)'': This histone modification is deposited by the polycomb complex PRC2. It is a clear marker of gene repression, and is likely bound by other proteins to exert a repressive function. Another polycomb complex, PRC1, can bindH3K27me3
H3K27me3 is an epigenetic modification to the DNA packaging protein histone H3. It is a mark that indicates the tri-methylation of lysine 27 on histone H3 protein.
This tri-methylation is associated with the Downregulation and upregulation, down ...
and adds the histone modification H2AK119Ub which aids chromatin compaction. Based on this data it appears that PRC1 is recruited through the action of PRC2, however, recent studies show that PRC1 is recruited to the same sites in the absence of PRC2.
;''Di and tri-methylation of H3 lysine 9 (H3K9me2/3)'': H3K9me2/3 is a well-characterised marker for heterochromatin
Heterochromatin is a tightly packed form of DNA or '' condensed DNA'', which comes in multiple varieties. These varieties lie on a continuum between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a rol ...
, and is therefore strongly associated with gene repression. The formation of heterochromatin has been best studied in the yeast ''Schizosaccharomyces pombe
''Schizosaccharomyces pombe'', also called "fission yeast", is a species of yeast used in traditional brewing and as a model organism in molecular and cell biology. It is a unicellular eukaryote, whose cells are rod-shaped. Cells typically meas ...
'', where it is initiated by recruitment of the RNA-induced transcriptional silencing (RITS) complex to double stranded RNAs produced from centromeric repeats. RITS recruits the Clr4 histone methyltransferase which deposits H3K9me2/3. This process is called histone methylation. H3K9Me2/3 serves as a binding site for the recruitment of Swi6 ( heterochromatin protein 1 or HP1, another classic heterochromatin marker) which in turn recruits further repressive activities including histone modifiers such as histone deacetylases and histone methyltransferases.
;''Trimethylation of H4 lysine 20 ( H4K20me3)'': This modification is tightly associated with heterochromatin, although its functional importance remains unclear. This mark is placed by the Suv4-20h methyltransferase, which is at least in part recruited by heterochromatin protein 1.
Bivalent promoters
Analysis of histone modifications in embryonic stem cells (and other stem cells) revealed many gene promoters carrying bothH3K4Me3
H3K4me3 is an epigenetic modification to the DNA packaging protein Histone H3 that indicates tri-methylation at the 4th lysine residue of the histone H3 protein and is often involved in the regulation of gene expression. The name denotes the addit ...
and H3K27Me3
H3K27me3 is an epigenetic modification to the DNA packaging protein histone H3. It is a mark that indicates the tri-methylation of lysine 27 on histone H3 protein.
This tri-methylation is associated with the Downregulation and upregulation, down ...
, in other words these promoters display both activating and repressing marks simultaneously. This peculiar combination of modifications marks genes that are poised for transcription; they are not required in stem cells, but are rapidly required after differentiation into some lineages. Once the cell starts to differentiate, these bivalent promoters are resolved to either active or repressive states depending on the chosen lineage.
Other functions
DNA damage repair
Marking sites of DNA damage is an important function for histone modifications. Without a repair marker, DNA would get destroyed by damage accumulated from sources such as theultraviolet radiation
Ultraviolet radiation, also known as simply UV, is electromagnetic radiation of wavelengths of 10–400 nanometers, shorter than that of visible light, but longer than X-rays. UV radiation is present in sunlight and constitutes about 10% of t ...
of the sun.
; ''Phosphorylation of H2AX at serine 139 (γH2AX)'': Phosphorylated H2AX (also known as gamma H2AX) is a marker for DNA double strand breaks, and forms part of the response to DNA damage. H2AX is phosphorylated early after detection of DNA double strand break, and forms a domain extending many kilobases either side of the damage. Gamma H2AX acts as a binding site for the protein MDC1, which in turn recruits key DNA repair proteins (this complex topic is well reviewed in) and as such, gamma H2AX forms a vital part of the machinery that ensures genome stability.
;''Acetylation of H3 lysine 56 (H3K56Ac)'': H3K56Acx is required for genome stability. H3K56 is acetylated by the p300/Rtt109 complex, but is rapidly deacetylated around sites of DNA damage. H3K56 acetylation is also required to stabilise stalled replication forks, preventing dangerous replication fork collapses. Although in general mammals make far greater use of histone modifications than microorganisms, a major role of H3K56Ac in DNA replication exists only in fungi, and this has become a target for antibiotic development.
; ''Trimethylation of H3 lysine 36 (H3K36me3)''
:H3K36me3 has the ability to recruit the MSH2-MSH6 (hMutSα) complex of the DNA mismatch repair
DNA mismatch repair (MMR) is a system for recognizing and repairing erroneous insertion, deletion, and mis-incorporation of nucleobase, bases that can arise during DNA replication and Genetic recombination, recombination, as well as DNA repair, ...
pathway. Consistently, regions of the human genome with high levels of H3K36me3 accumulate less somatic mutations due to mismatch repair
DNA mismatch repair (MMR) is a system for recognizing and repairing erroneous insertion, deletion, and mis-incorporation of nucleobase, bases that can arise during DNA replication and Genetic recombination, recombination, as well as DNA repair, ...
activity.
Chromosome condensation
; ''Phosphorylation of H3 at serine 10 (phospho-H3S10)'': The mitotic kinase aurora B phosphorylates histone H3 at serine 10, triggering a cascade of changes that mediate mitotic chromosome condensation. Condensed chromosomes therefore stain very strongly for this mark, but H3S10 phosphorylation is also present at certain chromosome sites outside mitosis, for example in pericentric heterochromatin of cells during G2. H3S10 phosphorylation has also been linked to DNA damage caused by R-loop formation at highly transcribed sites. ;''Phosphorylation H2B at serine 10/14 (phospho-H2BS10/14)'': Phosphorylation of H2B at serine 10 (yeast) or serine 14 (mammals) is also linked to chromatin condensation, but for the very different purpose of mediating chromosome condensation during apoptosis. This mark is not simply a late acting bystander in apoptosis as yeast carrying mutations of this residue are resistant to hydrogen peroxide-induced apoptotic cell death.Addiction
Epigenetic modifications of histone tails in specific regions of the brain are of central importance in addictions. Once particular epigenetic alterations occur, they appear to be long lasting "molecular scars" that may account for the persistence of addictions.Cigarette
A cigarette is a narrow cylinder containing a combustible material, typically tobacco, that is rolled into Rolling paper, thin paper for smoking. The cigarette is ignited at one end, causing it to smolder; the resulting smoke is orally inhale ...
smokers (about 15% of the US population) are usually addicted to nicotine
Nicotine is a natural product, naturally produced alkaloid in the nightshade family of plants (most predominantly in tobacco and ''Duboisia hopwoodii'') and is widely used recreational drug use, recreationally as a stimulant and anxiolytic. As ...
. After 7 days of nicotine treatment of mice, acetylation of both histone H3 and histone H4 was increased at the FosB promoter in the nucleus accumbens
The nucleus accumbens (NAc or NAcc; also known as the accumbens nucleus, or formerly as the ''nucleus accumbens septi'', Latin for ' nucleus adjacent to the septum') is a region in the basal forebrain rostral to the preoptic area of the hypo ...
of the brain, causing 61% increase in FosB expression. This would also increase expression of the splice variant
Alternative splicing, alternative RNA splicing, or differential splicing, is an alternative splicing process during gene expression that allows a single gene to produce different splice variants. For example, some exons of a gene may be included ...
Delta FosB. In the nucleus accumbens
The nucleus accumbens (NAc or NAcc; also known as the accumbens nucleus, or formerly as the ''nucleus accumbens septi'', Latin for ' nucleus adjacent to the septum') is a region in the basal forebrain rostral to the preoptic area of the hypo ...
of the brain, Delta FosB functions as a "sustained molecular switch" and "master control protein" in the development of an addiction
Addiction is a neuropsychological disorder characterized by a persistent and intense urge to use a drug or engage in a behavior that produces natural reward, despite substantial harm and other negative consequences. Repetitive drug use can ...
.
About 7% of the US population is addicted to alcohol
Alcohol may refer to:
Common uses
* Alcohol (chemistry), a class of compounds
* Ethanol, one of several alcohols, commonly known as alcohol in everyday life
** Alcohol (drug), intoxicant found in alcoholic beverages
** Alcoholic beverage, an alco ...
. In rats exposed to alcohol for up to 5 days, there was an increase in histone 3 lysine 9 acetylation in the pronociceptin promoter in the brain amygdala
The amygdala (; : amygdalae or amygdalas; also '; Latin from Greek language, Greek, , ', 'almond', 'tonsil') is a paired nucleus (neuroanatomy), nuclear complex present in the Cerebral hemisphere, cerebral hemispheres of vertebrates. It is c ...
complex. This acetylation is an activating mark for pronociceptin. The nociceptin/nociceptin opioid receptor system is involved in the reinforcing or conditioning effects of alcohol.
Methamphetamine
Methamphetamine (contracted from ) is a potent central nervous system (CNS) stimulant that is mainly used as a recreational drug use, recreational or Performance-enhancing substance, performance-enhancing drug and less commonly as a secon ...
addiction occurs in about 0.2% of the US population. Chronic methamphetamine use causes methylation of the lysine in position 4 of histone 3 located at the promoters of the '' c-fos'' and the '' C-C chemokine receptor 2 (ccr2)'' genes, activating those genes in the nucleus accumbens (NAc). c-fos is well known to be important in addiction
Addiction is a neuropsychological disorder characterized by a persistent and intense urge to use a drug or engage in a behavior that produces natural reward, despite substantial harm and other negative consequences. Repetitive drug use can ...
. The ''ccr2'' gene is also important in addiction, since mutational inactivation of this gene impairs addiction.
Histone Chaperones
Histone chaperones (biology) are specialized proteins that assist in the proper handling, transport, and assembly of histones, preventing their aggregation and ensuring their appropriate deposition onto DNA. These proteins play a crucial role in regulatingnucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone, histone proteins and resembles thread wrapped around a bobbin, spool. The nucleosome ...
assembly and disassembly, influencing transcriptional activity, DNA replication, and repair. Unlike enzymatic chromatin remodeling
Chromatin remodeling is the dynamic modification of chromatin architecture to allow access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby control gene expression. Such remodeling is principally carried out ...
, histone chaperones function by binding histones in a regulated manner, modulating chromatin structure without direct catalytic activity.
One key function of histone chaperones is maintaining a reservoir of histones, regulating their supply to ensure proper chromatin formation. During DNA replication and transcription (biology)
Transcription is the process of copying a segment of DNA into RNA for the purpose of gene expression. Some segments of DNA are transcribed into RNA molecules that can encode proteins, called messenger RNA (mRNA). Other segments of DNA are transc ...
, histone chaperones such as ASF1 and FACT facilitate nucleosome reassembly, ensuring the preservation of histone modifications that define cellular identity. Moreover, histone chaperones contribute to nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone, histone proteins and resembles thread wrapped around a bobbin, spool. The nucleosome ...
disassembly in response to cellular stress or DNA damage, thereby allowing access to repair machinery.
Histone chaperones also participate in the selective deposition of histone variants, which are functionally distinct from canonical histones. For example, HIRA is a chaperone that specifically deposits the histone variant H3.3, a marker of active chromatin regions. Similarly, CAF-1 is responsible for incorporating H3.1 and H3.2 into newly replicated DNA, highlighting the functional specialization within chaperone networks.
Given their critical roles, misregulation of histone chaperones has been implicated in diseases such as cancer. Aberrant chaperone activity can lead to improper histone deposition, genome instability
Genome instability (also genetic instability or genomic instability) refers to a high frequency of mutations within the genome of a cellular lineage. These mutations can include changes in nucleic acid sequences, chromosomal rearrangements or ...
, and altered gene expression, contributing to tumorigenesis
Carcinogenesis, also called oncogenesis or tumorigenesis, is the formation of a cancer, whereby normal cells are transformed into cancer cells. The process is characterized by changes at the cellular, genetic, and epigenetic levels and abn ...
. Current research is exploring histone chaperones as potential therapeutic targets, particularly in cancers characterized by disrupted chromatin landscapes.
Chaperone Networks
The coordinated action of multiple histone chaperones forms an intricate network responsible for histone transport, Chromatin assembly factor 1, andgenome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as ...
maintenance. Chaperone networks facilitate the transport of histones which are synthesized in the cytoplasm
The cytoplasm describes all the material within a eukaryotic or prokaryotic cell, enclosed by the cell membrane, including the organelles and excluding the nucleus in eukaryotic cells. The material inside the nucleus of a eukaryotic cell a ...
and must be escorted to the cell nucleus
The cell nucleus (; : nuclei) is a membrane-bound organelle found in eukaryote, eukaryotic cell (biology), cells. Eukaryotic cells usually have a single nucleus, but a few cell types, such as mammalian red blood cells, have #Anucleated_cells, ...
. This network ensures histones are deposited at the appropriate genomic locations, maintaining chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important r ...
integrity and function.
Histone chaperones play a crucial role in responding to DNA damage by regulating chromatin accessibility. For example, in response to double strand breaks, chaperones such as FACT and ASF1 help disassemble nucleosomes at damage sites, allowing repair factors to access the lesion. Once repair is completed, these chaperones facilitate the reassembly of nucleosomes, restoring chromatin structure and ensuring epigenetic information is maintained.
In addition to their role in genome stability, histone chaperones contribute to epigenetic inheritance. During cell division, chromatin states must be faithfully propagated to daughter cells. Chaperones help distribute parental histones onto newly synthesized DNA strands, preserving histone modifications and ensuring continuity of cellular identity. Disruptions in these processes can lead to epigenetic abnormalities associated with developmental disorders.
Synthesis
The first step of chromatin structure duplication is the synthesis of histone proteins: H1, H2A, H2B, H3, H4. These proteins are synthesized during S phase of the cell cycle. There are different mechanisms which contribute to the increase of histone synthesis.Yeast
Yeast carry one or two copies of each histone gene, which are not clustered but rather scattered throughout chromosomes. Histone gene transcription is controlled by multiple gene regulatory proteins such as transcription factors which bind to histone promoter regions. In budding yeast, the candidate gene for activation of histone gene expression is SBF. SBF is a transcription factor that is activated in late G1 phase, when it dissociates from its repressor Whi5. This occurs when Whi5 is phosphorylated by Cdc8 which is a G1/S Cdk. Suppression of histone gene expression outside of S phases is dependent on Hir proteins which form inactive chromatin structure at the locus of histone genes, causing transcriptional activators to be blocked.Metazoan
Inmetazoan
Animals are multicellular, eukaryotic organisms in the biological kingdom Animalia (). With few exceptions, animals consume organic material, breathe oxygen, have myocytes and are able to move, can reproduce sexually, and grow from a ho ...
s the increase in the rate of histone synthesis is due to the increase in processing of pre-mRNA to its mature form as well as decrease in mRNA degradation; this results in an increase of active mRNA for translation of histone proteins. The mechanism for mRNA activation has been found to be the removal of a segment of the 3' end of the mRNA strand, and is dependent on association with stem-loop binding protein ( SLBP). SLBP also stabilizes histone mRNAs during S phase by blocking degradation by the 3'hExo nuclease. SLBP levels are controlled by cell-cycle proteins, causing SLBP to accumulate as cells enter S phase and degrade as cells leave S phase. SLBP are marked for degradation by phosphorylation at two threonine residues by cyclin dependent kinases, possibly cyclin A/ cdk2, at the end of S phase. Metazoans also have multiple copies of histone genes clustered on chromosomes which are localized in structures called Cajal bodies as determined by genome-wide chromosome conformation capture analysis (4C-Seq).
Link between cell-cycle control and synthesis
Nuclear protein Ataxia-Telangiectasia (NPAT), also known as nuclear protein coactivator of histone transcription, is a transcription factor which activates histone gene transcription on chromosomes 1 and 6 of human cells. NPAT is also a substrate of cyclin E-Cdk2, which is required for the transition between G1 phase and S phase. NPAT activates histone gene expression only after it has been phosphorylated by the G1/S-Cdk cyclin E-Cdk2 in early S phase. This shows an important regulatory link between cell-cycle control and histone synthesis.History
Histones were discovered in 1884 byAlbrecht Kossel
Ludwig Karl Martin Leonhard Albrecht Kossel (; 16 September 1853 – 5 July 1927) was a biochemist and pioneer in the study of genetics. He was awarded the Nobel Prize for Physiology or Medicine in 1910 for his work in determining the chemical ...
. The word "histone" dates from the late 19th century and is derived from the German word ''"Histon"'', a word itself of uncertain origin, perhaps from Ancient Greek ''ἵστημι'' (hístēmi, “make stand”) or ''ἱστός'' (histós, “loom”).
In the early 1960s, before the types of histones were known and before histones were known to be highly conserved across taxonomically diverse organisms, James F. Bonner and his collaborators began a study of these proteins that were known to be tightly associated with the DNA in the nucleus of higher organisms. Bonner and his postdoctoral fellow Ru Chih C. Huang showed that isolated chromatin would not support RNA transcription in the test tube, but if the histones were extracted from the chromatin, RNA could be transcribed from the remaining DNA. Their paper became a citation classic. Paul T'so and James Bonner had called together a World Congress on Histone Chemistry and Biology in 1964, in which it became clear that there was no consensus on the number of kinds of histone and that no one knew how they would compare when isolated from different organisms.James Bonner and Paul T'so (1965) ''The Nucleohistones''. Holden-Day Inc, San Francisco, London, Amsterdam. Bonner and his collaborators then developed methods to separate each type of histone, purified individual histones, compared amino acid compositions in the same histone from different organisms, and compared amino acid sequences of the same histone from different organisms in collaboration with Emil Smith from UCLA. For example, they found Histone IV sequence to be highly conserved between peas and calf thymus. However, their work on the biochemical characteristics of individual histones did not reveal how the histones interacted with each other or with DNA to which they were tightly bound.
Also in the 1960s, Vincent Allfrey and Alfred Mirsky had suggested, based on their analyses of histones, that acetylation and methylation of histones could provide a transcriptional control mechanism, but did not have available the kind of detailed analysis that later investigators were able to conduct to show how such regulation could be gene-specific. Until the early 1990s, histones were dismissed by most as inert packing material for eukaryotic nuclear DNA, a view based in part on the models of Mark Ptashne and others, who believed that transcription was activated by protein-DNA and protein-protein interactions on largely naked DNA templates, as is the case in bacteria.
During the 1980s, Yahli Lorch and Roger Kornberg showed that a nucleosome on a core promoter prevents the initiation of transcription in vitro, and Michael Grunstein demonstrated that histones repress transcription in vivo, leading to the idea of the nucleosome as a general gene repressor. Relief from repression is believed to involve both histone modification and the action of chromatin-remodeling complexes. Vincent Allfrey and Alfred Mirsky had earlier proposed a role of histone modification in transcriptional activation, regarded as a molecular manifestation of epigenetics. Michael Grunstein and David Allis found support for this proposal, in the importance of histone acetylation for transcription in yeast and the activity of the transcriptional activator Gcn5 as a histone acetyltransferase.
The discovery of the H5 histone appears to date back to the 1970s, and it is now considered an isoform
A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene and are the result of genetic differences. While many perform the same or similar biological roles, some isoforms have uniqu ...
of Histone H1.
See also
* Histone variants *Chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important r ...
* Gene silencing
Gene silencing is the regulation of gene expression in a cell to prevent the expression of a certain gene. Gene silencing can occur during either Transcription (genetics), transcription or Translation (biology), translation and is often used in res ...
* Genetics
Genetics is the study of genes, genetic variation, and heredity in organisms.Hartl D, Jones E (2005) It is an important branch in biology because heredity is vital to organisms' evolution. Gregor Mendel, a Moravian Augustinians, Augustinian ...
* Histone acetyltransferase
Histone acetyltransferases (HATs) are enzymes that acetylation, acetylate conserved lysine amino acids on histone proteins by transferring an acetyl group from acetyl-CoA to form ε-N-acetyllysine, ε-''N''-acetyllysine. DNA is wrapped around his ...
* Histone deacetylases
* Histone methyltransferase
* Histone-modifying enzymes
* Nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone, histone proteins and resembles thread wrapped around a bobbin, spool. The nucleosome ...
* PRMT4 pathway
* Protamine
* Histone H1
References
External links
HistoneDB 2.0 - Database of histones and variants
at
NCBI
The National Center for Biotechnology Information (NCBI) is part of the National Library of Medicine (NLM), a branch of the National Institutes of Health (NIH). It is approved and funded by the government of the United States. The NCBI is loca ...
Chromatin, Histones & Cathepsin
PMAP The Proteolysis Map-animation {{Authority control Epigenetics Proteins DNA-binding proteins