|
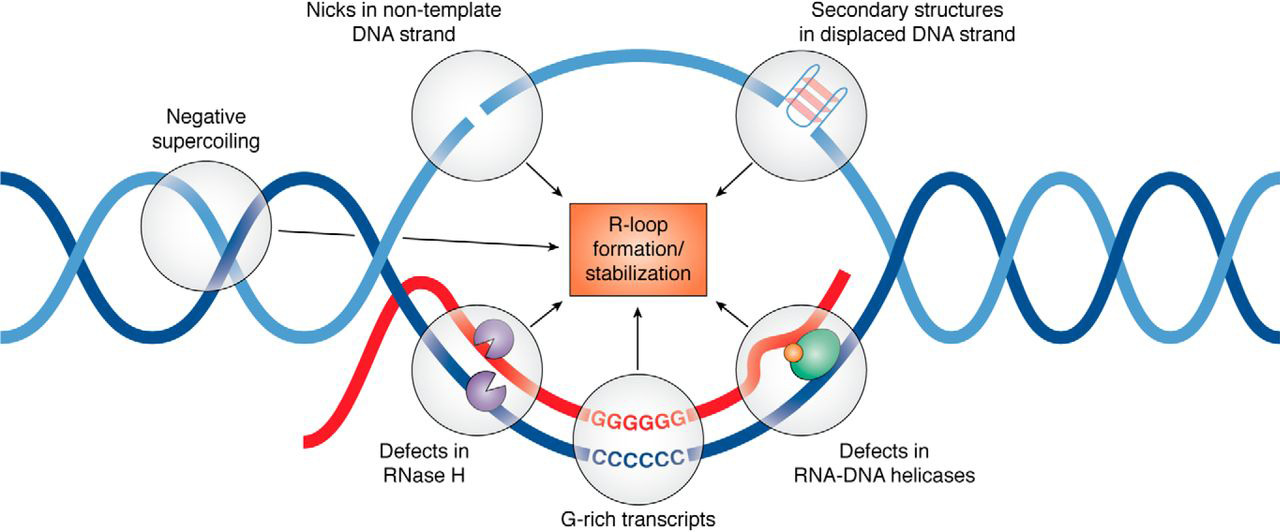
R-loop
An R-loop is a three-stranded nucleic acid structure, composed of a DNA:RNA hybrid and the associated non-template single-stranded DNA. R-loops may be formed in a variety of circumstances, and may be tolerated or cleared by cellular components. The term "R-loop" was given to reflect the similarity of these structures to D-loops; the "R" in this case represents the involvement of an RNA moiety. In the laboratory, R-loops may also be created by the hybridization of mature mRNA with double-stranded DNA under conditions favoring the formation of a DNA-RNA hybrid; in this case, the intron regions (which have been spliced out of the mRNA) form single-stranded DNA loops, as they cannot hybridize with complementary sequence in the mRNA. History R-looping was first described in 1976. Independent R-looping studies from the laboratories of Richard J. Roberts and Phillip A. Sharp showed that protein coding adenovirus genes contained DNA sequences that were not present in the mature ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DRIP-seq
DRIP-seq (DRIP-sequencing) is a technology for genome-wide profiling of a type of DNA-RNA hybrid called an "R-loop". DRIP-seq utilizes a sequence-independent but structure-specific antibody for DNA-RNA immunoprecipitation (DRIP) to capture R-loops for massively parallel DNA sequencing. Introduction An R-loop is a three-stranded nucleic acid structure, which consists of a DNA-RNA hybrid duplex and a displaced single stranded DNA (ssDNA). R-loops are predominantly formed in cytosine-rich genomic regions during transcription and are known to be involved with gene expression and immunoglobulin class switching. They have been found in a variety of species, ranging from bacteria to mammals. They are preferentially localized at CpG island promoters in human cells and highly transcribed regions in yeast. Under abnormal conditions, namely elevated production of DNA-RNA hybrids, R-loops can cause genome instability by exposing single-stranded DNA to endogenous damages exerted by the acti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intron
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e. a region inside a gene."The notion of the cistron .e., gene... must be replaced by that of a transcription unit containing regions which will be lost from the mature messenger – which I suggest we call introns (for intragenic regions) – alternating with regions which will be expressed – exons." (Gilbert 1978) The term ''intron'' refers to both the DNA sequence within a gene and the corresponding RNA sequence in RNA transcripts. The non-intron sequences that become joined by this RNA processing to form the mature RNA are called exons. Introns are found in the genes of most organisms and many viruses and they can be located in both protein-coding genes and genes that function as RNA (noncoding genes). There are four main types of introns: tRNA introns, group I introns, group II introns, and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
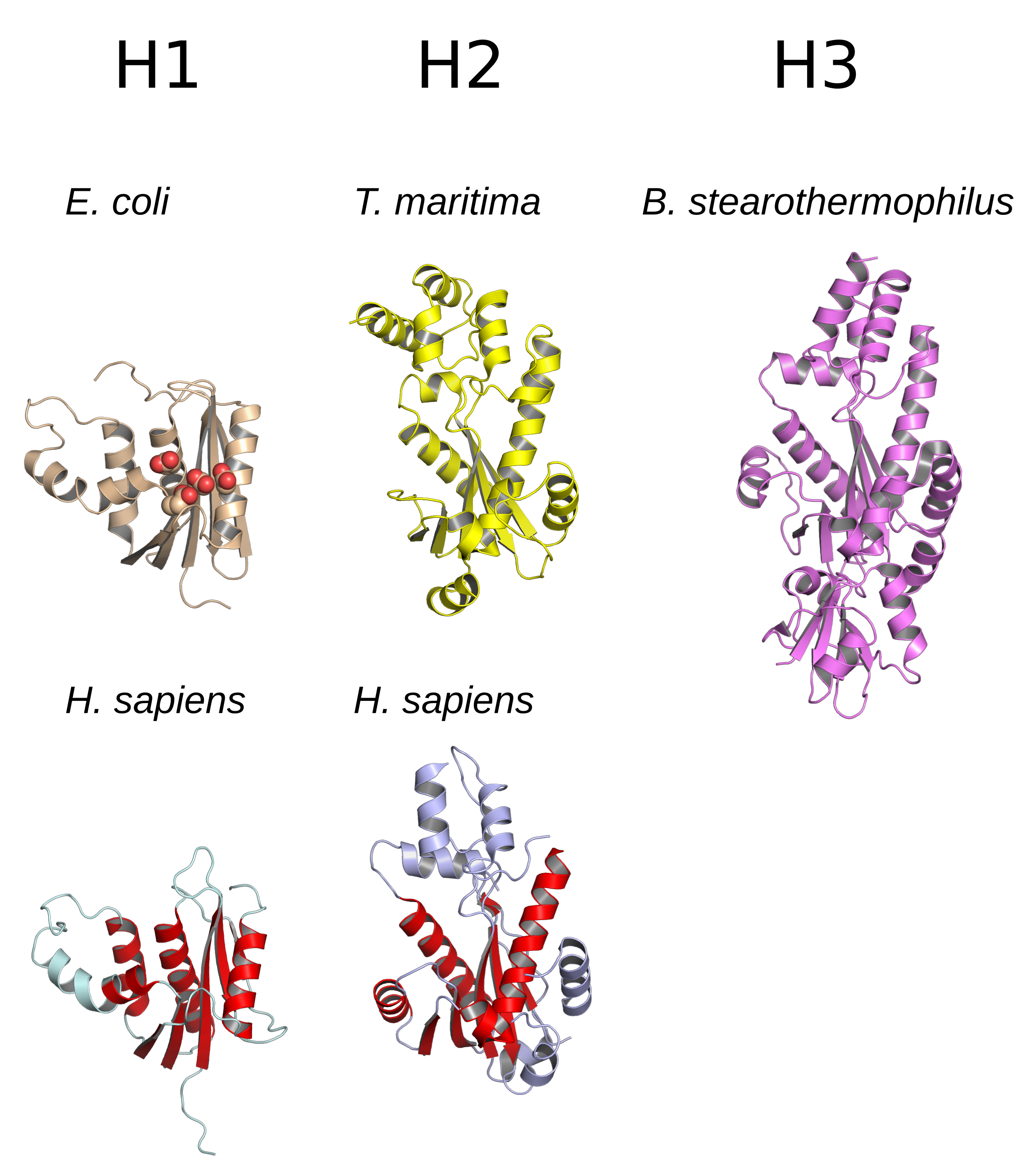
Ribonuclease H
Ribonuclease H (abbreviated RNase H or RNH) is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/ DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes. The family is divided into evolutionarily related groups with slightly different substrate preferences, broadly designated ribonuclease H1 and H2. The human genome encodes both H1 and H2. Human ribonuclease H2 is a heterotrimeric complex composed of three subunits, mutations in any of which are among the genetic causes of a rare disease known as Aicardi–Goutières syndrome. A third type, closely related to H2, is found only in a few prokaryotes, whereas H1 and H2 occur in all domains of life. Additionally, RNase H1-like retroviral ribonuclease H domains occur in multidomain reverse transcriptase proteins, which are encoded by retroviruses such as HIV and are required for viral repli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
D-loop
In molecular biology, a displacement loop or D-loop is a DNA structure where the two strands of a double-stranded DNA molecule are separated for a stretch and held apart by a third strand of DNA. An R-loop is similar to a D-loop, but in this case the third strand is RNA rather than DNA. The third strand has a base sequence which is complementary to one of the main strands and pairs with it, thus displacing the other complementary main strand in the region. Within that region the structure is thus a form of triple-stranded DNA. A diagram in the paper introducing the term illustrated the D-loop with a shape resembling a capital "D", where the displaced strand formed the loop of the "D". D-loops occur in a number of particular situations, including in DNA repair, in telomeres, and as a semi-stable structure in mitochondrial circular DNA molecules. In mitochondria Researchers at Caltech discovered in 1971 that the circular mitochondrial DNA from growing cells included a sh ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methylation
In the chemical sciences, methylation denotes the addition of a methyl group on a substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These terms are commonly used in chemistry, biochemistry, soil science, and the biological sciences. In biological systems, methylation is catalyzed by enzymes; such methylation can be involved in modification of heavy metals, regulation of gene expression, regulation of protein function, and RNA processing. In vitro methylation of tissue samples is also one method for reducing certain histological staining artifacts. The reverse of methylation is demethylation. In biology In biological systems, methylation is accomplished by enzymes. Methylation can modify heavy metals, regulate gene expression, RNA processing and protein function. It has been recognized as a key process underlying epigenetics. Methanogenesis Methanogenesis, the proce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Promoter (genetics)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Promoters control gene expression in bacteria and eukaryotes. RNA polymerase must attach to DNA near a gene for transcription to occur. Promoter DNA sequences provide an enzyme binding site. The -10 sequence is TATAAT. -35 sequences are conserved on average, but not in most promoters. Artificial promoters with conserved -10 and -35 elements transcribe more slowly. All ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
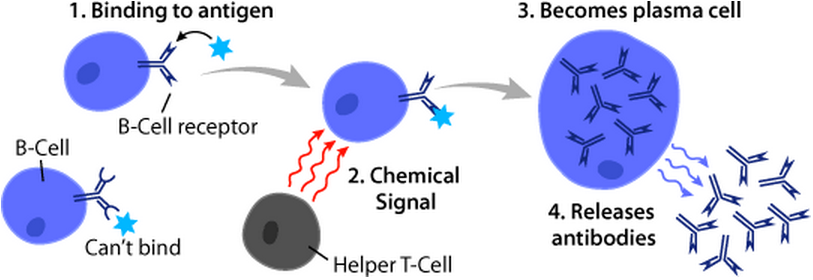
B Cell
B cells, also known as B lymphocytes, are a type of white blood cell of the lymphocyte subtype. They function in the humoral immunity component of the adaptive immune system. B cells produce antibody molecules which may be either secreted or inserted into the plasma membrane where they serve as a part of B-cell receptors. When a naïve or memory B cell is activated by an antigen, it proliferates and differentiates into an antibody-secreting effector cell, known as a plasmablast or plasma cell. Additionally, B cells Antigen presentation, present antigens (they are also classified as professional Antigen-presenting cell, antigen-presenting cells (APCs)) and secrete cytokines. In mammals, B cells Cellular differentiation, mature in the bone marrow, which is at the core of most bones. In birds, B cells mature in the bursa of Fabricius, a lymphoid organ where they were first discovered by Chang and Glick, which is why the 'B' stands for bursa and not bone marrow as commonly believed ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Immunoglobulin Class Switching
Immunoglobulin class switching, also known as isotype switching, isotypic commutation or class-switch recombination (CSR), is a biological mechanism that changes a B cell's production of immunoglobulin from one type to another, such as from the isotype IgM to the isotype IgG. During this process, the constant-region portion of the antibody heavy chain is changed, but the variable region of the heavy chain stays the same (the terms ''variable'' and ''constant'' refer to changes or lack thereof between antibodies that target different epitopes). Since the variable region does not change, class switching does not affect antigen specificity. Instead, the antibody retains affinity for the same antigens, but can interact with different effector molecules. Mechanism Class switching occurs after activation of a mature B cell via its membrane-bound antibody molecule (or B cell receptor) to generate the different classes of antibody, all with the same variable domains as the origi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequencing
In genetics and biochemistry, sequencing means to determine the primary structure (sometimes incorrectly called the primary sequence) of an unbranched biopolymer. Sequencing results in a symbolic linear depiction known as a sequence which succinctly summarizes much of the atomic-level structure of the sequenced molecule. DNA sequencing DNA sequencing is the process of determining the nucleotide order of a given DNA fragment. So far, most DNA sequencing has been performed using the chain termination method developed by Frederick Sanger. This technique uses sequence-specific termination of a DNA synthesis reaction using modified nucleotide substrates. However, new sequencing technologies such as pyrosequencing are gaining an increasing share of the sequencing market. More genome data are now being produced by pyrosequencing than Sanger DNA sequencing. Pyrosequencing has enabled rapid genome sequencing. Bacterial genomes can be sequenced in a single run with several times cove ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endogeny (biology)
Endogenous substances and processes are those that originate from within a living system such as an organism, tissue, or cell. In contrast, exogenous substances and processes are those that originate from outside of an organism. For example, estradiol is an endogenous estrogen hormone produced within the body, whereas ethinylestradiol is an exogenous synthetic estrogen, commonly used in birth control pills Oral contraceptives, abbreviated OCPs, also known as birth control pills, are medications taken by mouth for the purpose of birth control. Female Two types of female oral contraceptive pill, taken once per day, are widely available: * The combi .... References External links *{{Wiktionary-inline, endogeny Biology ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Topoisomerase
DNA topoisomerases (or topoisomerases) are enzymes that catalyze changes in the topological state of DNA, interconverting relaxed and supercoiled forms, linked (catenated) and unlinked species, and knotted and unknotted DNA. Topological issues in DNA arise due to the intertwined nature of its double-helical structure, which, for example, can lead to overwinding of the DNA duplex during DNA replication and transcription. If left unchanged, this torsion would eventually stop the DNA or RNA polymerases involved in these processes from continuing along the DNA helix. A second topological challenge results from the linking or tangling of DNA during replication. Left unresolved, links between replicated DNA will impede cell division. The DNA topoisomerases prevent and correct these types of topological problems. They do this by binding to DNA and cutting the sugar-phosphate backbone of either one (type I topoisomerases) or both (type II topoisomerases) of the DNA strands. This transien ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |