|
D-loop
In molecular biology, a displacement loop or D-loop is a DNA structure where the two strands of a double-stranded DNA molecule are separated for a stretch and held apart by a third strand of DNA. An R-loop is similar to a D-loop, but in that case the third strand is RNA rather than DNA. The third strand has a base sequence which is complementary to one of the main strands and pairs with it, thus displacing the other complementary main strand in the region. Within that region the structure is thus a form of triple-stranded DNA. A diagram in the paper introducing the term illustrated the D-loop with a shape resembling a capital "D", where the displaced strand formed the loop of the "D". D-loops occur in a number of particular situations, including in DNA repair, in telomeres, and as a semi-stable structure in mitochondrial circular DNA molecules. In mitochondria Researchers at Caltech discovered in 1971 that the circular mitochondrial DNA from growing cells included a sho ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MtDNA Control Region
A mitochondrion is a specialized organelle found in the cytoplasm of eukaryotic cells, which is the powerhouse of cells that produce energy through oxidative phosphorylation. Besides producing energy, they are crucial for various cellular functions such as calcium signaling, controlling metabolism, synthesizing hemoglobin and steroids, and regulating programmed cell death. Besides the production of ATP, there is a complex relationship between the mitochondrial genome (mtDNA) and the nuclear genome (nDNA). Unlike nuclear DNA which resides in the nucleus, mtDNA is inherited from the mother. Upon fertilization, the mtDNA of the sperm is typically lost through ubiquitination, and only the egg contributes to the mtDNA of the zygote, due to the significant difference in mtDNA copy number between the sperm (about 100 copies) and the egg (about 100,000 copies). Mitochondrial DNA (mtDNA) Is a small, abundant, and purified molecule of DNA. It is a closed, circular, double-stranded molecu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homologous Recombination
Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids (usually DNA as in Cell (biology), cellular organisms but may be also RNA in viruses). Homologous recombination is widely used by cells to accurately DNA repair, repair harmful DNA breaks that occur on both strands of DNA, known as double-strand breaks (DSB), in a process called homologous recombinational repair (HRR). Homologous recombination also produces new combinations of DNA sequences during meiosis, the process by which eukaryotes make gamete cells, like sperm and ovum, egg cells in animals. These new combinations of DNA represent genetic variation in offspring, which in turn enables populations to Adaptation, adapt during the course of evolution. Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Shelterin
Shelterin (also called telosome) is a protein complex known to protect telomeres in many eukaryotes from DNA repair mechanisms, as well as to regulate telomerase activity. In mammals and other vertebrates, telomeric DNA consists of repeating double-stranded 5'-TTAGGG-3' (G-strand) sequences (2-15 kilobases in humans) along with the 3'-AATCCC-5' (C-strand) complement, ending with a 50-400 nucleotide 3' (G-strand) overhang. Much of the final double-stranded portion of the telomere forms a T-loop (Telomere-loop) that is invaded by the 3' (G-strand) overhang to form a small D-loop (Displacement-loop). The absence of shelterin causes telomere uncapping and thereby activates damage-signaling pathways that may lead to non-homologous end joining (NHEJ), homology directed repair (HDR), end-to-end fusions, genomic instability, senescence, or apoptosis. Subunits Shelterin has six subunits: TRF1, TRF2, POT1, RAP1, TIN2, and TPP1. They can operate in smaller subsets to r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Triple-stranded DNA
Triple-stranded DNA (also known as H-DNA or Triplex-DNA) is a DNA structure in which three oligonucleotides wind around each other and form a triple helix. In triple-stranded DNA, the third strand binds to a Nucleic acid double helix#Helix geometries, B-form DNA (via Base pair, Watson–Crick base-pairing) double helix by forming Hoogsteen base pairs or reversed Hoogsteen hydrogen bonds. Structure Examples of triple-stranded DNA from natural sources with the necessary combination of base composition and structural elements have been described, for example in Satellite DNA. Hoogsteen base pairing A thymine (T) nucleobase can bind to a Base pair, Watson–Crick base-pairing of T-A by forming a Hoogsteen base pair, Hoogsteen hydrogen bond. The thymine hydrogen bonds with the adenosine (A) of the original double-stranded DNA to create a T-A*T base-triplet. Intermolecular and intramolecular interactions There are two classes of triplex DNA: intermolecular and intramolecula ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
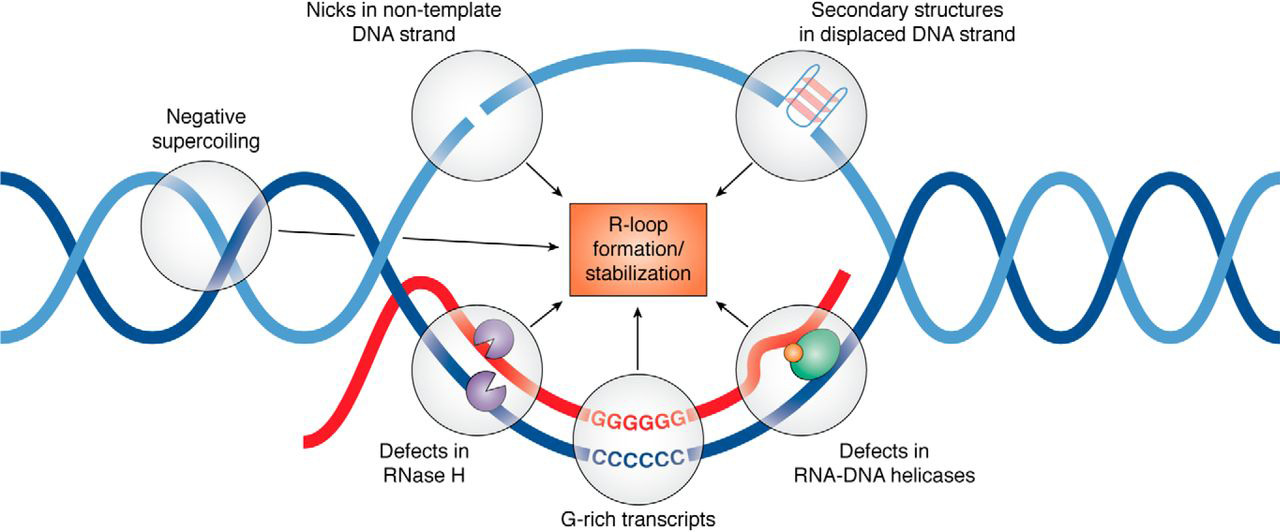
R-loop
An R-loop is a three-stranded nucleic acid structure, composed of a DNA:RNA hybrid and the associated non-template single-stranded DNA. R-loops may be formed in a variety of circumstances and may be tolerated or cleared by cellular components. The term "R-loop" was given to reflect the similarity of these structures to D-loops; the "R" in this case represents the involvement of an RNA moiety (chemistry), moiety. In the laboratory, R-loops can be created by transcription of DNA sequences (for example those that have a high GC content) that favor annealing of the RNA behind the progressing RNA polymerase. At least 100bp of DNA:RNA hybrid is required to form a stable R-loop structure. R-loops may also be created by the Nucleic acid hybridization#Hybridization, hybridization of mature mRNA with double-stranded DNA under conditions favoring the formation of a DNA-RNA hybrid; in this case, the intron regions (which have been Splicing (genetics), spliced out of the mRNA) form single-stra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is constantly modified in Cell (biology), cells, by internal metabolism, metabolic by-products, and by external ionizing radiation, ultraviolet light, and medicines, resulting in spontaneous DNA damage involving tens of thousands of individual molecular lesions per cell per day. DNA modifications can also be programmed. Molecular lesions can cause structural damage to the DNA molecule, and can alter or eliminate the cell's ability for Transcription (biology), transcription and gene expression. Other lesions may induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells following mitosis. Consequently, DNA repair as part of the DNA damage response (DDR) is constantly active. When normal repair proce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Telomere
A telomere (; ) is a region of repetitive nucleotide sequences associated with specialized proteins at the ends of linear chromosomes (see #Sequences, Sequences). Telomeres are a widespread genetic feature most commonly found in eukaryotes. In most, if not all species possessing them, they protect the terminal regions of DNA, chromosomal DNA from progressive degradation and ensure the integrity of linear chromosomes by preventing DNA repair systems from mistaking the very ends of the DNA strand for a double-strand break. Discovery The existence of a special structure at the ends of chromosomes was independently proposed in 1938 by Hermann Joseph Muller, studying the fruit fly ''Drosophila melanogaster'', and in 1939 by Barbara McClintock, working with maize. Muller observed that the ends of irradiated fruit fly chromosomes did not present alterations such as deletions or inversions. He hypothesized the presence of a protective cap, which he coined "telomeres", from the Greek ' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all life, living organisms, acting as the most essential part of heredity, biological inheritance. This is essential for cell division during growth and repair of damaged tissues, while it also ensures that each of the new cells receives its own copy of the DNA. The cell possesses the distinctive property of division, which makes replication of DNA essential. DNA is made up of a nucleic acid double helix, double helix of two Complementary DNA, complementary DNA strand, strands. DNA is often called double helix. The double helix describes the appearance of a double-stranded DNA which is composed of two linear strands that run opposite to each other and twist together. During replication, these strands are separated. Each strand of the original DNA molecule then serves as a template for the production of its counterpart, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mitochondrial DNA
Mitochondrial DNA (mtDNA and mDNA) is the DNA located in the mitochondrion, mitochondria organelles in a eukaryotic cell that converts chemical energy from food into adenosine triphosphate (ATP). Mitochondrial DNA is a small portion of the DNA contained in a eukaryotic cell; most of the DNA is in the cell nucleus, and, in plants and algae, the DNA also is found in plastids, such as chloroplasts. Mitochondrial DNA is responsible for coding of 13 essential subunits of the complex oxidative phosphorylation (OXPHOS) system which has a role in cellular energy conversion. Human mitochondrial DNA was the first significant part of the human genome to be sequenced. This sequencing revealed that human mtDNA has 16,569 base pairs and encodes 13 proteins. As in other vertebrates, the human mitochondrial genetic code differs slightly from nuclear DNA. Since animal mtDNA evolves faster than nuclear genetic markers, it represents a mainstay of phylogenetics and evolutionary biology. It als ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genes (journal)
''Genes'' is a quarterly peer-reviewed open access scientific journal that is published by MDPI. The editor-in-chief is J. Peter W. Young (University of York). It covers all topics related to genes, genetics, and genomics. Abstracting and indexing The journal is abstracted and indexed in: * Chemical Abstracts * EBSCOhost * EMBASE * Science Citation Index Expanded * Scopus Scopus is a scientific abstract and citation database, launched by the academic publisher Elsevier as a competitor to older Web of Science in 2004. The ensuing competition between the two databases has been characterized as "intense" and is c ... References External links * Genetics journals Open access journals Academic journals established in 2010 Quarterly journals English-language journals MDPI academic journals {{genetics-journal-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
POT1
Protection of telomeres protein 1 is a protein that in humans is encoded by the ''POT1'' gene. Function This gene is a member of the telombin family and encodes a nuclear protein involved in telomere maintenance. Specifically, this protein functions as a member of a multi-protein complex known as shelterin, that binds to the TTAGGG repeats of telomeres, regulating telomere length and protecting chromosome ends from illegitimate recombination, catastrophic chromosome instability, and abnormal chromosome segregation. Alternatively spliced transcript variants have been described. The absence of POT1 in mouse embryonic fibroblasts and chicken cells leads to a detrimental DNA damage response on telomeres resulting in telomere dysfunction-induced foci (TIFs). POT1 is required for telomere protection because it allows for telomere inhibition of DNA damage response factors. The protein also serves a role in the regulation of telomerase activity on telomeres. In vitro experiments util ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RAD51
DNA repair protein RAD51 homolog 1 is a protein encoded by the gene ''RAD51''. The enzyme encoded by this gene is a member of the RAD51 protein family which assists in repair of DNA double strand breaks. RAD51 family members are homologous to the bacterial RecA, Archaeal RadA, and yeast Rad51. The protein is highly conserved in most eukaryotes, from yeast to humans. The name RAD51 derives from ''radiation sensitive protein 51''. Variants Two alternatively spliced transcript variants of this gene have been reported, which encode distinct proteins. Transcript variants utilizing alternative polyA signals also exist. Family In mammals, seven recA-like genes have been identified: Rad51, Rad51L1/B, Rad51L2/C, Rad51L3/D, XRCC2, XRCC3, and DMC1/Lim15. All of these proteins, with the exception of meiosis-specific DMC1, are essential for development in mammals. Rad51 is a member of thRecA-like NTPases Function In humans, RAD51 is a 339-amino acid protein that plays ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |