|
MtDNA Control Region
The mtDNA control region is an area of the mitochondrial genome which is non-coding DNA. This region controls RNA and DNA synthesis. It is the most polymorphic region of the human mtDNA genome, with polymorphism concentrated in hypervariable regions. The average nucleotide diversity in these regions is 1.7%. Despite this variability, an RNA transcript from this region has a conserved secondary structure (''pictured'') which has been found to be under selective pressure. The mtDNA control region contains the origin of replication of one strand, and the origin of transcription for both strands. There is also an open reading frame thought to code for 7s ribosomal RNA in humans but not in mice or cows, where it has been deleted. Distinction from D-loop The control region and mtDNA D-loop are sometimes used synonymously in the literature; specifically the control region includes the D-loop along with adjacent transcription promoter regions. For this reason, the control region ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the three dimensional form of ''local segments'' of proteins. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein folds into its three dimensional tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the amino hydrogen and carboxyl oxygen atoms in the peptide backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik Linderstrøm-Lang at Stanford in 1952. Other types of biopolymers such as nucleic acids also possess characteristic secondary structures. Types The most common secondary st ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ribosomal RNA
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal DNA (rDNA) and then bound to ribosomal proteins to form small and large ribosome subunits. rRNA is the physical and mechanical factor of the ribosome that forces transfer RNA (tRNA) and messenger RNA (mRNA) to process and translate the latter into proteins. Ribosomal RNA is the predominant form of RNA found in most cells; it makes up about 80% of cellular RNA despite never being translated into proteins itself. Ribosomes are composed of approximately 60% rRNA and 40% ribosomal proteins by mass. Structure Although the primary structure of rRNA sequences can vary across organisms, base-pairing within these sequences commonly forms stem-loop configurations. The length and position of these rRNA stem-loops allow them to create three-dimensio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
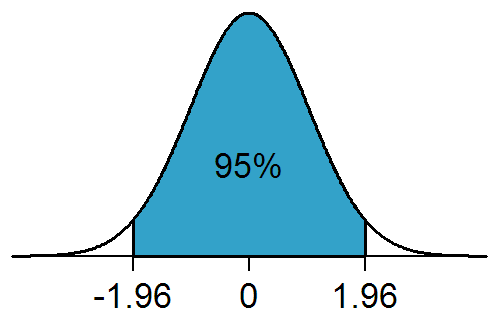
Statistical Significance
In statistical hypothesis testing, a result has statistical significance when it is very unlikely to have occurred given the null hypothesis (simply by chance alone). More precisely, a study's defined significance level, denoted by \alpha, is the probability of the study rejecting the null hypothesis, given that the null hypothesis is true; and the ''p''-value of a result, ''p'', is the probability of obtaining a result at least as extreme, given that the null hypothesis is true. The result is statistically significant, by the standards of the study, when p \le \alpha. The significance level for a study is chosen before data collection, and is typically set to 5% or much lower—depending on the field of study. In any experiment or observation that involves drawing a sample from a population, there is always the possibility that an observed effect would have occurred due to sampling error alone. But if the ''p''-value of an observed effect is less than (or equal to) the significan ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endurance Training
Endurance training is the act of exercising to increase endurance. The term endurance training generally refers to training the aerobic system as opposed to the anaerobic system. The need for endurance in sports is often predicated as the need of cardiovascular and simple muscular endurance, but the issue of endurance is far more complex. Endurance can be divided into two categories including: general endurance and specific endurance. It can be shown that endurance in sport is closely tied to the execution of skill and technique. A well conditioned athlete can be defined as, the athlete who executes his or her technique consistently and effectively with the least effort. Key for measuring endurance are heart rate, power in cycling and pace in running. Endurance in sports Endurance training is essential for a variety of endurance sports. A notable example is distance running events (800 meters upwards to marathon and ultra-marathon) with the required degree of endurance trainin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
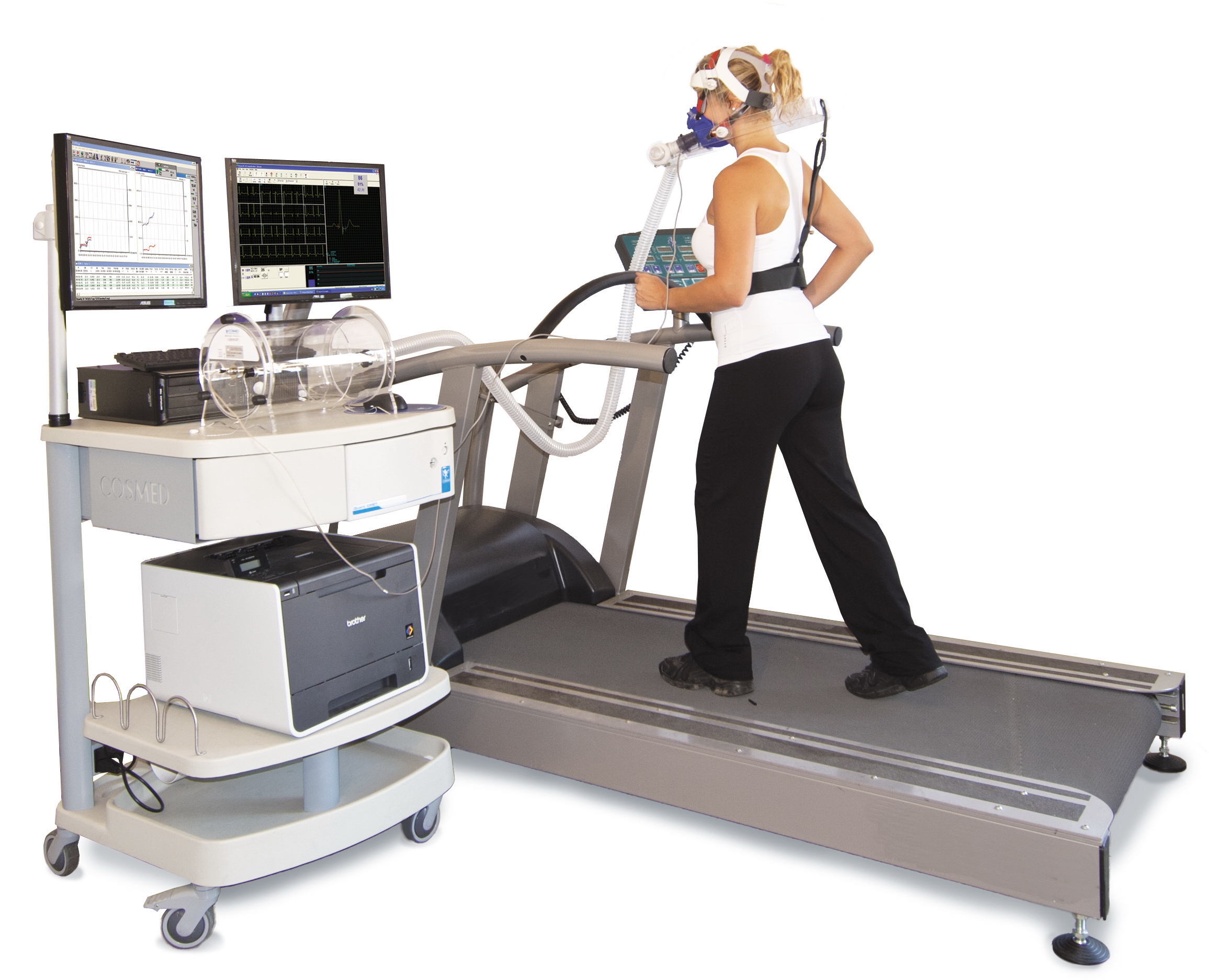
VO2 Max
VO2 max (also maximal oxygen consumption, maximal oxygen uptake or maximal aerobic capacity) is the maximum rate of oxygen consumption attainable during physical exertion. The name is derived from three abbreviations: "V̇" for volume (the dot appears over the V to indicate "per unit of time"), "O2" for oxygen, and "max" for maximum. A similar measure is VO2 peak (peak oxygen consumption), which is the measurable value from a session of physical exercise, be it incremental or otherwise. It could match or underestimate the actual VO2 max. Confusion between the values in older and popular fitness literature is common. The measurement of V̇O2 max in the laboratory provides a quantitative value of endurance fitness for comparison of individual training effects and between people in endurance training. Maximal oxygen consumption reflects cardiorespiratory fitness and endurance capacity in exercise performance. Elite athletes, such as competitive distance runners, racing cyclists o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery. Knowledge of DNA sequences has become indispensable for basic biological research, DNA Genographic Projects and in numerous applied fields such as medical diagnosis, biotechnology, forensic biology, virology and biological systematics. Comparing healthy and mutated DNA sequences can diagnose different diseases including various cancers, characterize antibody repertoire, and can be used to guide patient treatment. Having a quick way to sequence DNA allows for faster and more individualized medical care to be administered, and for more organisms to be identified and cataloged. The rapid speed of sequencing attained with m ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endurance
Endurance (also related to sufferance, resilience, constitution, fortitude, and hardiness) is the ability of an organism to exert itself and remain active for a long period of time, as well as its ability to resist, withstand, recover from and have immunity to trauma, wounds or fatigue. It is usually used in aerobic or anaerobic exercise. The definition of 'long' varies according to the type of exertion – minutes for high intensity anaerobic exercise, hours or days for low intensity aerobic exercise. Training for endurance can reduce the ability to exert endurance strength unless an individual also undertakes resistance training to counteract this effect. When a person is able to accomplish or withstand a higher amount of effort than their original capabilities their endurance is increasing which to many personnel indicates progress. In looking to improve one's endurance they may slowly increase the amount of repetitions or time spent, if higher repetitions are taken ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Acronym
An acronym is a word or name formed from the initial components of a longer name or phrase. Acronyms are usually formed from the initial letters of words, as in '' NATO'' (''North Atlantic Treaty Organization''), but sometimes use syllables, as in '' Benelux'' (short for ''Belgium, the Netherlands, and Luxembourg''). They can also be a mixture, as in '' radar'' (''Radio Detection And Ranging''). Acronyms can be pronounced as words, like '' NASA'' and '' UNESCO''; as individual letters, like ''FBI'', '' TNT'', and ''ATM''; or as both letters and words, like ''JPEG'' (pronounced ') and ''IUPAC''. Some are not universally pronounced one way or the other and it depends on the speaker's preference or the context in which it is being used, such as '' SQL'' (either "sequel" or "ess-cue-el"). The broader sense of ''acronym''—the meaning of which includes terms pronounced as letters—is sometimes criticized, but it is the term's original meaning and is in common use. Dictionary ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Promoter (biology)
In genetics, a promoter is a sequence of DNA to which proteins bind to initiate transcription of a single RNA transcript from the DNA downstream of the promoter. The RNA transcript may encode a protein (mRNA), or can have a function in and of itself, such as tRNA or rRNA. Promoters are located near the transcription start sites of genes, upstream on the DNA (towards the 5' region of the sense strand). Promoters can be about 100–1000 base pairs long, the sequence of which is highly dependent on the gene and product of transcription, type or class of RNA polymerase recruited to the site, and species of organism. Promoters control gene expression in bacteria and eukaryotes. RNA polymerase must attach to DNA near a gene for transcription to occur. Promoter DNA sequences provide an enzyme binding site. The -10 sequence is TATAAT. -35 sequences are conserved on average, but not in most promoters. Artificial promoters with conserved -10 and -35 elements transcribe more slowly. All ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
D-loop
In molecular biology, a displacement loop or D-loop is a DNA structure where the two strands of a double-stranded DNA molecule are separated for a stretch and held apart by a third strand of DNA. An R-loop is similar to a D-loop, but in this case the third strand is RNA rather than DNA. The third strand has a base sequence which is complementary to one of the main strands and pairs with it, thus displacing the other complementary main strand in the region. Within that region the structure is thus a form of triple-stranded DNA. A diagram in the paper introducing the term illustrated the D-loop with a shape resembling a capital "D", where the displaced strand formed the loop of the "D". D-loops occur in a number of particular situations, including in DNA repair, in telomeres, and as a semi-stable structure in mitochondrial circular DNA molecules. In mitochondria Researchers at Caltech discovered in 1971 that the circular mitochondrial DNA from growing cells included a sh ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Deletion (genetics)
In genetics, a deletion (also called gene deletion, deficiency, or deletion mutation) (sign: Δ) is a mutation (a genetic aberration) in which a part of a chromosome or a sequence of DNA is left out during DNA replication. Any number of nucleotides can be deleted, from a single base to an entire piece of chromosome. Some chromosomes have fragile spots where breaks occur which result in the deletion of a part of chromosome. The breaks can be induced by heat, viruses, radiations, chemicals. When a chromosome breaks, a part of it is deleted or lost, the missing piece of chromosome is referred to as deletion or a deficiency. For synapsis to occur between a chromosome with a large intercalary deficiency and a normal complete homolog, the unpaired region of the normal homolog must loop out of the linear structure into a deletion or compensation loop. The smallest single base deletion mutations occur by a single base flipping in the template DNA, followed by template DNA strand slip ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Open Reading Frame
In molecular biology, open reading frames (ORFs) are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a Prokaryote, prokaryotic DNA sequence, where only one of the #Six-frame translation, six possible reading frames will be "open" (the "reading", however, refers to the RNA produced by Transcription (biology), transcription of the DNA and its subsequent interaction with the ribosome in Translation (biology), translation). Such an ORF may contain a start codon (usually AUG in terms of RNA) and by definition cannot extend beyond a stop codon (usually UAA, UAG or UGA in RNA). That start codon (not necessarily the first) indicates where translation may start. The transcription terminator, transcription termination site is located after the ORF, beyond the Translation (biology), translation stop codon. If transcription were to cease before the stop codon, an incomplete protein would be made during translation. In ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |