R-loop on:
[Wikipedia]
[Google]
[Amazon]
 An R-loop is a three-stranded nucleic acid structure, composed of a DNA:
An R-loop is a three-stranded nucleic acid structure, composed of a DNA:
 R-looping was first described in 1976. Independent R-looping studies from the laboratories of Richard J. Roberts and Phillip A. Sharp showed that
R-looping was first described in 1976. Independent R-looping studies from the laboratories of Richard J. Roberts and Phillip A. Sharp showed that
 An R-loop is a three-stranded nucleic acid structure, composed of a DNA:
An R-loop is a three-stranded nucleic acid structure, composed of a DNA:RNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA and deoxyrib ...
hybrid and the associated non-template single-stranded DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
. R-loops may be formed in a variety of circumstances and may be tolerated or cleared by cellular components. The term "R-loop" was given to reflect the similarity of these structures to D-loop
In molecular biology, a displacement loop or D-loop is a DNA structure where the two strands of a double-stranded DNA molecule are separated for a stretch and held apart by a third strand of DNA. An R-loop is similar to a D-loop, but in that cas ...
s; the "R" in this case represents the involvement of an RNA moiety.
In the laboratory, R-loops can be created by transcription of DNA sequences (for example those that have a high GC content) that favor annealing of the RNA behind the progressing RNA polymerase. At least 100bp of DNA:RNA hybrid is required to form a stable R-loop structure. R-loops may also be created by the hybridization of mature mRNA with double-stranded DNA under conditions favoring the formation of a DNA-RNA hybrid; in this case, the intron
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e., a region inside a gene."The notion of the cistron .e., gen ...
regions (which have been spliced out of the mRNA) form single-stranded DNA loops, as they cannot hybridize with complementary sequence in the mRNA.
History
 R-looping was first described in 1976. Independent R-looping studies from the laboratories of Richard J. Roberts and Phillip A. Sharp showed that
R-looping was first described in 1976. Independent R-looping studies from the laboratories of Richard J. Roberts and Phillip A. Sharp showed that protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
coding adenovirus
Adenoviruses (members of the family ''Adenoviridae'') are medium-sized (90–100 nm), nonenveloped (without an outer lipid bilayer) viruses with an icosahedral nucleocapsid containing a double-stranded DNA genome. Their name derives from t ...
gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei ...
s contained DNA sequences that were not present in the mature mRNA. Roberts and Sharp were awarded the Nobel Prize
The Nobel Prizes ( ; ; ) are awards administered by the Nobel Foundation and granted in accordance with the principle of "for the greatest benefit to humankind". The prizes were first awarded in 1901, marking the fifth anniversary of Alfred N ...
in 1993 for independently discovering introns. After their discovery in adenovirus, introns were found in a number of eukaryotic
The eukaryotes ( ) constitute the Domain (biology), domain of Eukaryota or Eukarya, organisms whose Cell (biology), cells have a membrane-bound cell nucleus, nucleus. All animals, plants, Fungus, fungi, seaweeds, and many unicellular organisms ...
genes such as the eukaryotic ovalbumin
Ovalbumin (abbreviated OVA) is the main protein found in egg white, making up approximately 55% of the total protein. Ovalbumin displays sequence and three-dimensional homology to the serpin superfamily, but unlike most serpins it is not a serine ...
gene (first by the O'Malley laboratory, then confirmed by other groups), hexon DNA, and extrachromosomal rRNA
Ribosomal ribonucleic acid (rRNA) is a type of non-coding RNA which is the primary component of ribosomes, essential to all cells. rRNA is a ribozyme which carries out protein synthesis in ribosomes. Ribosomal RNA is transcribed from ribosomal ...
genes of '' Tetrahymena thermophila''.
In the mid-1980s, development of an antibody
An antibody (Ab) or immunoglobulin (Ig) is a large, Y-shaped protein belonging to the immunoglobulin superfamily which is used by the immune system to identify and neutralize antigens such as pathogenic bacteria, bacteria and viruses, includin ...
that binds specifically to the R-loop structure opened the door for immunofluorescence
Immunofluorescence (IF) is a light microscopy-based technique that allows detection and localization of a wide variety of target biomolecules within a cell or tissue at a quantitative level. The technique utilizes the binding specificity of anti ...
studies, as well as genome-wide characterization of R-loop formation by DRIP-seq.
R-loop mapping
R-loop mapping is a laboratory technique used to distinguish introns fromexon
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequence ...
s in double-stranded DNA. These R-loops are visualized by electron microscopy
An electron microscope is a microscope that uses a beam of electrons as a source of illumination. It uses electron optics that are analogous to the glass lenses of an optical light microscope to control the electron beam, for instance focusing i ...
and reveal intron regions of DNA by creating unbound loops at these regions.
R-loops ''in vivo''
The potential for R-loops to serve as replication primers was demonstrated in 1980. In 1994, R-loops were demonstrated to be present ''in vivo'' through analysis of plasmids isolated from ''E. coli
''Escherichia coli'' ( )Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. is a gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus ''Escherichia'' that is commonly foun ...
'' mutants carrying mutations in topoisomerase
DNA topoisomerases (or topoisomerases) are enzymes that catalyze changes in the topological state of DNA, interconverting relaxed and supercoiled forms, linked (catenated) and unlinked species, and knotted and unknotted DNA. Topological issues in ...
. This discovery of endogenous
Endogeny, in biology, refers to the property of originating or developing from within an organism, tissue, or cell.
For example, ''endogenous substances'', and ''endogenous processes'' are those that originate within a living system (e.g. an ...
R-loops, in conjunction with rapid advances in genetic sequencing
In genetics and biochemistry, sequencing means to determine the primary structure (sometimes incorrectly called the primary sequence) of an unbranched biopolymer. Sequencing results in a symbolic linear depiction known as a sequence which succ ...
technologies, inspired a blossoming of R-loop research in the early 2000s that continues to this day.
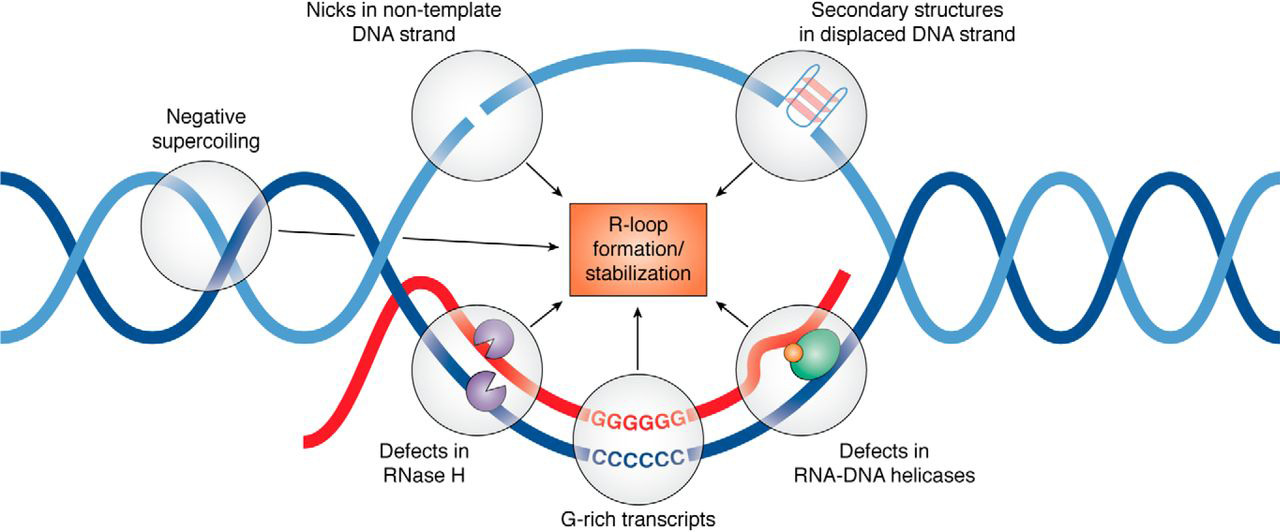
Regulation of R-loop formation and resolution
More than 50 proteins that appear to influence R-loop accumulation, and while many of them are believed to contribute by sequestering or processing newly transcribed RNA to prevent re-annealing to the template, mechanisms of R-loop interaction for many of these proteins remain to be determined. There are three main classes of enzyme that can remove RNA that becomes trapped in the duplex within an R-loop. RNaseH enzymes are the primary proteins responsible for the dissolution of R-loops, acting to degrade the RNA moiety in order to allow the two complementary DNA strands to anneal. Alternatively, Helicases act to unwind the RNA:DNA duplex so that RNA is released. Senataxin is one helicase that can move along ssRNA, and appears to be necessary for preventing R-loop formation at transcription pause sites. The third enzyme class capable of removing R-loops are branchpoint translocases such as FANCM, SMARCAL1 and ZRANB3 in humans or RecG in bacteria. Branchpoint translocases act on the double-stranded DNA adjacent to the DNA:RNA hybrid. By pushing at the branchpoint, they act to "zip up" the DNA and expel the trapped RNA. This makes branchpoint translocases efficient at removing both RNA and proteins that are bound to the R-loop structure. Branchpoint translocases may work together with RNaseH and helicases on some types of R-loops that occur at challenging structures.Roles of R-loops in genetic regulation
R-loop formation is a key step inimmunoglobulin class switching
Immunoglobulin class switching, also known as isotype switching, isotypic commutation or class-switch recombination (CSR), is a biological mechanism that changes a B cell's production of immunoglobulin from one type to another, such as from the ...
, a process that allows activated B cell
B cells, also known as B lymphocytes, are a type of the lymphocyte subtype. They function in the humoral immunity component of the adaptive immune system. B cells produce antibody molecules which may be either secreted or inserted into the plasm ...
s to modulate antibody
An antibody (Ab) or immunoglobulin (Ig) is a large, Y-shaped protein belonging to the immunoglobulin superfamily which is used by the immune system to identify and neutralize antigens such as pathogenic bacteria, bacteria and viruses, includin ...
production. They also appear to play a role in protecting some active promoters from methylation
Methylation, in the chemistry, chemical sciences, is the addition of a methyl group on a substrate (chemistry), substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replac ...
. The presence of R-loops can also inhibit transcription. Additionally, R-loop formation appears to be associated with “open” chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important r ...
, characteristic of actively transcribed regions.
R-loops as genetic damage
When unscheduled R-loops form, they can cause damage by a number of different mechanisms. Exposed single-strandedDNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
can come under attack by endogenous mutagens, including DNA-modifying enzymes such as activation-induced cytidine deaminase
Activation-induced cytidine deaminase, also known as AICDA, AID and single-stranded DNA cytosine deaminase, is a 24 kDa enzyme which in humans is encoded by the ''AICDA'' gene. It creates mutations in DNA by deamination of cytosine base, which ...
, and can block replication forks to induce fork collapse and subsequent double-strand breaks. As well, R-loops may induce unscheduled replication by acting as a primer.
R-loop accumulation has been associated with a number of diseases, including amyotrophic lateral sclerosis type 4 (ALS4), ataxia oculomotor apraxia type 2 (AOA2), Aicardi–Goutières syndrome, Angelman syndrome
Angelman syndrome (AS) is a genetic disorder that affects approximately 1 in 15,000 individuals. AS impairs the function of the nervous system, producing symptoms, such as severe intellectual disability, developmental disability, limited to no ...
, Prader–Willi syndrome
Prader–Willi syndrome (PWS) is a rare genetic disorder caused by a loss of function of specific genes on chromosome 15. In newborns, symptoms include hypotonia, weak muscles, poor feeding, and slow development. Beginning in childhood, those ...
, and cancer. Genes associated with Fanconi anemia
Fanconi anemia (FA) is a rare, autosomal recessive genetic disease characterized by aplastic anemia, congenital defects, endocrinological abnormalities, and an increased incidence of developing cancer. The study of Fanconi anemia has improve ...
also seem to be important for the maintenance of genome stability under conditions where R-loops accumulate.
R-loops, Introns and DNA damage
Intron
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e., a region inside a gene."The notion of the cistron .e., gen ...
s are non-coding regions within gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei ...
s that are transcribed along with the coding regions of genes, but are subsequently removed from the primary RNA transcript by splicing. Actively transcribed regions of DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
often form R-loops that are vulnerable to DNA damage
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is constantly modified ...
. Introns reduce R-loop formation and DNA damage in highly expressed yeast genes. Genome-wide analysis showed that intron-containing genes display decreased R-loop levels and decreased DNA damage compared to intron-less genes of similar expression in both yeast and humans. Inserting an intron within an R-loop prone gene can also suppress R-loop formation and recombination. Bonnet et al. (2017) speculated that the function of introns in maintaining genetic stability may explain their evolutionary maintenance at certain locations, particularly in highly expressed genes.
See also
* DRIP-seq *Ribonuclease H
Ribonuclease H (abbreviated RNase H or RNH) is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly al ...
* Immunoglobulin class switching
Immunoglobulin class switching, also known as isotype switching, isotypic commutation or class-switch recombination (CSR), is a biological mechanism that changes a B cell's production of immunoglobulin from one type to another, such as from the ...
* DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all life, living organisms, acting as the most essential part of heredity, biolog ...
References
{{reflist DNA RNA splicing