
In
biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the
DNA sequence
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. Th ...
.
The
Greek prefix ''
epi-'' ( "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in addition to" the traditional
genetic basis for inheritance.
Epigenetics most often involves changes that affect the
regulation of gene expression
Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products (protein or RNA). Sophisticated programs of gene expression are wide ...
, but the term can also be used to describe any heritable phenotypic change. Such effects on
cellular
Cellular may refer to:
*Cellular automaton, a model in discrete mathematics
* Cell biology, the evaluation of cells work and more
* ''Cellular'' (film), a 2004 movie
*Cellular frequencies, assigned to networks operating in cellular RF bands
*Cell ...
and
physiological
Physiology (; ) is the scientific study of functions and mechanisms in a living system. As a sub-discipline of biology, physiology focuses on how organisms, organ systems, individual organs, cells, and biomolecules carry out the chemical ...
phenotypic trait
A phenotypic trait, simply trait, or character state is a distinct variant of a phenotypic characteristic of an organism; it may be either inherited or determined environmentally, but typically occurs as a combination of the two.Lawrence, Eleano ...
s may result from external or
environmental
A biophysical environment is a biotic and abiotic surrounding of an organism or population, and consequently includes the factors that have an influence in their survival, development, and evolution. A biophysical environment can vary in scale f ...
factors, or be part of normal development. It can also lead to diseases such as cancer.
The term also refers to the mechanism of changes: functionally relevant alterations to the
genome that do not involve mutation of the
nucleotide sequence. Examples of mechanisms that produce such changes are
DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts t ...
and
histone modification
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn ar ...
, each of which alters how genes are expressed without altering the underlying
DNA sequence. Gene expression can be controlled through the action of
repressor protein
In molecular genetics, a repressor is a DNA- or RNA-binding protein that inhibits the expression of one or more genes by binding to the operator or associated silencers. A DNA-binding repressor blocks the attachment of RNA polymerase to the ...
s that attach to
silencer regions of the DNA. These epigenetic changes may last through
cell divisions for the duration of the cell's life, and may also last for multiple generations, even though they do not involve changes in the underlying DNA sequence of the organism;
instead, non-genetic factors cause the organism's genes to behave (or "express themselves") differently.
One example of an epigenetic change in
eukaryotic biology is the process of
cellular differentiation
Cellular differentiation is the process in which a stem cell alters from one type to a differentiated one. Usually, the cell changes to a more specialized type. Differentiation happens multiple times during the development of a multicellular ...
. During
morphogenesis,
totipotent stem cells become the various
pluripotent cell line
An immortalised cell line is a population of cells from a multicellular organism which would normally not proliferate indefinitely but, due to mutation, have evaded normal cellular senescence and instead can keep undergoing division. The cell ...
s of the
embryo
An embryo is an initial stage of development of a multicellular organism. In organisms that reproduce sexually, embryonic development is the part of the life cycle that begins just after fertilization of the female egg cell by the male spe ...
, which in turn become fully differentiated cells. In other words, as a single fertilized
egg cell
The egg cell, or ovum (plural ova), is the female reproductive cell, or gamete, in most anisogamous organisms (organisms that reproduce sexually with a larger, female gamete and a smaller, male one). The term is used when the female gamete is ...
– the
zygote – continues to
divide, the resulting daughter cells change into all the different cell types in an organism, including
neurons
A neuron, neurone, or nerve cell is an electrically excitable cell that communicates with other cells via specialized connections called synapses. The neuron is the main component of nervous tissue in all animals except sponges and placozoa. N ...
,
muscle cells,
epithelium
Epithelium or epithelial tissue is one of the four basic types of animal tissue, along with connective tissue, muscle tissue and nervous tissue. It is a thin, continuous, protective layer of compactly packed cells with a little intercellul ...
,
endothelium of
blood vessels
The blood vessels are the components of the circulatory system that transport blood throughout the human body. These vessels transport blood cells, nutrients, and oxygen to the tissues of the body. They also take waste and carbon dioxide away f ...
, etc., by activating some genes while inhibiting the expression of others.
Definitions
The term ''epigenetics'' in its contemporary usage emerged in the 1990s, but for some years has been used with somewhat variable meanings.
A definition of the concept of ''epigenetic trait'' as a "stably heritable phenotype resulting from changes in a chromosome without alterations in the DNA sequence" was formulated at a
Cold Spring Harbor
Cold Spring Harbor is a hamlet and census-designated place (CDP) in the Town of Huntington, in Suffolk County, on the North Shore of Long Island in New York. As of the 2010 United States census, the CDP population was 5,070.
History
Cold Spri ...
meeting in 2008,
although alternate definitions that include non-heritable traits are still being used widely.
The term ''epigenesis'' has a generic meaning of "extra growth" that has been used in English since the 17th century.
Waddington's canalisation, 1940s
The hypothesis of epigenetic changes affecting the expression of
chromosomes was put forth by the Russian biologist
Nikolai Koltsov. From the generic meaning, and the associated adjective ''epigenetic'', British embryologist
C. H. Waddington
Conrad Hal Waddington (8 November 1905 – 26 September 1975) was a British developmental biologist, paleontologist, geneticist, embryologist and philosopher who laid the foundations for systems biology, epigenetics, and evolutionary developme ...
coined the term ''epigenetics'' in 1942 as pertaining to ''
epigenesis'', in parallel to
Valentin Haecker's 'phenogenetics' ().
[
"For the purpose of a study of inheritance, the relation between phenotypes and genotypes ..is, from a wider biological point of view, of crucial importance, since it is the kernel of the whole problem of development. Many geneticists have recognized this and attempted to discover the processes involved in the mechanism by which the genes of the genotype bring about phenotypic effects. The first step in such an enterprise is – or rather should be, since it is often omitted by those with undue respect for the powers of reason – to describe what can be seen of the developmental processes. For enquiries of this kind, the word 'phenogenetics' was coined by Haecker 918, The second and more important part of the task is to discover the causal mechanisms at work and to relate them as far as possible to what experimental embryology has already revealed of the mechanics of development. We might use the name 'epigenetics' for such studies, thus emphasizing their relation to the concepts, so strongly favourable to the classical theory of epigenesis, which have been reached by the experimental embryologists. We certainly need to remember that between genotype and phenotype, and connecting them to each other, there lies a whole complex of developmental processes. It is convenient to have a name for this complex: 'epigenotype' seems suitable."
] ''Epigenesis'' in the context of the biology of that period referred to the
differentiation of cells from their initial
totipotent state during
embryonic development
An embryo is an initial stage of development of a multicellular organism. In organisms that reproduce sexually, embryonic development is the part of the life cycle that begins just after fertilization of the female egg cell by the male sperm ...
.
When Waddington coined the term, the physical nature of
genes and their role in heredity was not known. He used it instead as a conceptual model of how genetic components might interact with their surroundings to produce a
phenotype; he used the phrase "
epigenetic landscape
In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the DNA sequence. The Greek prefix '' epi-'' ( "over, outside of, around") in ''epigenetics'' implies features that are "o ...
" as a metaphor for
biological development
Developmental biology is the study of the process by which animals and plants grow and develop. Developmental biology also encompasses the biology of regeneration, asexual reproduction, metamorphosis, and the growth and differentiation of stem ce ...
. Waddington held that cell fates were established during development in a process he called
canalisation much as a marble rolls down to the point of
lowest local elevation.
Waddington suggested visualising increasing irreversibility of cell type differentiation as ridges rising between the valleys where the marbles (analogous to cells) are travelling.
In recent times, Waddington's notion of the epigenetic landscape has been rigorously formalized in the context of the
systems dynamics
System dynamics (SD) is an approach to understanding the nonlinear behaviour of complex systems over time using stocks, flows, internal feedback loops, table functions and time delays.
Overview
System dynamics is a methodology and mathematical ...
state approach to the study of cell-fate.
Cell-fate determination is predicted to exhibit certain dynamics, such as attractor-convergence (the attractor can be an equilibrium point, limit cycle or
strange attractor
In the mathematical field of dynamical systems, an attractor is a set of states toward which a system tends to evolve, for a wide variety of starting conditions of the system. System values that get close enough to the attractor values remain ...
) or oscillatory.
Contemporary
Robin Holliday defined in 1990 epigenetics as "the study of the mechanisms of temporal and spatial control of gene activity during the development of complex organisms."
Thus, in its broadest sense, ''epigenetic'' can be used to describe anything other than DNA sequence that influences the development of an organism.
More recent usage of the word in biology follows stricter definitions. As defined by
Arthur Riggs and colleagues, it is "the study of
mitotically and/or
meiotically heritable changes in gene function that cannot be explained by changes in DNA sequence."
The term has also been used, however, to describe processes which have not been demonstrated to be heritable, such as some forms of histone modification. Consequently, there are attempts to redefine "epigenetics" in broader terms that would avoid the constraints of requiring
heritability
Heritability is a statistic used in the fields of breeding and genetics that estimates the degree of ''variation'' in a phenotypic trait in a population that is due to genetic variation between individuals in that population. The concept of h ...
. For example,
Adrian Bird
Sir Adrian Peter Bird, (born 3 July 1947) is a British geneticist and Buchanan Professor of Genetics at the University of Edinburgh. Bird has spent much of his academic career in Edinburgh, from receiving his PhD in 1970 to working at the MRC ...
defined epigenetics as "the structural adaptation of chromosomal regions so as to register, signal or perpetuate altered activity states."
This definition would be inclusive of transient modifications associated with
DNA repair or
cell-cycle phases as well as stable changes maintained across multiple cell generations, but exclude others such as templating of membrane architecture and
prions unless they impinge on chromosome function. Such redefinitions however are not universally accepted and are still subject to debate.
The
NIH "Roadmap Epigenomics Project", ongoing as of 2016, uses the following definition: "For purposes of this program, epigenetics refers to both heritable changes in gene activity and
expression (in the progeny of cells or of individuals) and also stable, long-term alterations in the transcriptional potential of a cell that are not necessarily heritable."
In 2008, a consensus definition of the epigenetic trait, a "stably heritable phenotype resulting from changes in a chromosome without alterations in the DNA sequence," was made at a
Cold Spring Harbor
Cold Spring Harbor is a hamlet and census-designated place (CDP) in the Town of Huntington, in Suffolk County, on the North Shore of Long Island in New York. As of the 2010 United States census, the CDP population was 5,070.
History
Cold Spri ...
meeting.
The similarity of the word to "genetics" has generated many parallel usages. The "
epigenome" is a parallel to the word "
genome," referring to the overall epigenetic state of a cell, and
epigenomics refers to global analyses of epigenetic changes across the entire genome.
The phrase "
genetic code" has also been adapted – the "
epigenetic code The epigenetic code is hypothesised to be a defining code in every eukaryotic cell consisting of the specific epigenetic modification in each cell. It consists of histone modifications defined by the histone code and additional epigenetic modificati ...
" has been used to describe the set of epigenetic features that create different phenotypes in different cells from the same underlying DNA sequence. Taken to its extreme, the "epigenetic code" could represent the total state of the cell, with the position of each molecule accounted for in an ''epigenomic map'', a diagrammatic representation of the gene expression, DNA methylation and histone modification status of a particular genomic region. More typically, the term is used in reference to systematic efforts to measure specific, relevant forms of epigenetic information such as the
histone code
The histone code is a hypothesis that the transcription of genetic information encoded in DNA is in part regulated by chemical modifications (known as ''histone marks'') to histone proteins, primarily on their unstructured ends. Together with sim ...
or
DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts t ...
patterns.
Developmental psychology
In a sense somewhat unrelated to its use in any biological disciplines, the term "epigenetic" has also been used in
developmental psychology
Developmental psychology is the science, scientific study of how and why humans grow, change, and adapt across the course of their lives. Originally concerned with infants and children, the field has expanded to include adolescence, adult deve ...
to describe psychological development as the result of an ongoing, bi-directional interchange between heredity and the environment. Interactive ideas of development have been discussed in various forms and under various names throughout the 19th and 20th centuries. An early version was proposed, among the founding statements in
embryology, by
Karl Ernst von Baer and popularized by
Ernst Haeckel
Ernst Heinrich Philipp August Haeckel (; 16 February 1834 – 9 August 1919) was a German zoologist, naturalist, eugenicist, philosopher, physician, professor, marine biologist and artist. He discovered, described and named thousands of new sp ...
. A radical epigenetic view, known as physiological epigenesis, was developed by
Paul Wintrebert Paul Wintrebert (1867–1966) was a French embryologist and a theoretician of developmental biology.
He coined the term cytoskeleton (''cytosquelette'') in 1931.
He held radical epigenetic views. In his 60s, he published a trilogy in which he d ...
. Another variation, probabilistic epigenesis, was presented by
Gilbert Gottlieb in 2003. This view encompasses all of the possible developing factors on an organism and how they not only influence the organism and each other but how the organism also influences its own development. Gottlieb gave an example of Rhesus monkeys where infants that did not receive typical maternal care lacked
serotonin
Serotonin () or 5-hydroxytryptamine (5-HT) is a monoamine neurotransmitter. Its biological function is complex and multifaceted, modulating mood, cognition, reward, learning, memory, and numerous physiological processes such as vomiting and vas ...
, which in turn made them more aggressive as they got older. On another note, the long-standing notion "cells that fire together, wire together" derives from
Hebbian theory which asserts that
synaptogenesis, a developmental process with great epigenetic precedence, depends on the activity of the respective synapses within a neural network. Where experience alters the excitability of neurons, increased neural activity has been linked to increased demethylation .
The developmental psychologist
Erik Erikson
Erik Homburger Erikson (born Erik Salomonsen; 15 June 1902 – 12 May 1994) was a German-American developmental psychologist and psychoanalyst known for his theory on psychological development of human beings. He coined the phrase identity cr ...
wrote of an ''epigenetic principle'' in his 1968 book
''Identity: Youth and Crisis'', encompassing the notion that we develop through an unfolding of our personality in predetermined stages, and that our environment and surrounding culture influence how we progress through these stages. This biological unfolding in relation to our socio-cultural settings is done in
stages of psychosocial development, where "progress through each stage is in part determined by our success, or lack of success, in all the previous stages."
Although empirical studies have yielded discrepant results, epigenetic modifications are thought to be a biological mechanism for
transgenerational trauma.
Molecular basis
Epigenetic changes modify the activation of certain genes, but not the genetic code sequence of DNA. The microstructure (not code) of DNA itself or the associated
chromatin proteins may be modified, causing activation or silencing. This mechanism enables differentiated cells in a multicellular organism to express only the genes that are necessary for their own activity. Epigenetic changes are preserved when cells divide. Most epigenetic changes only occur within the course of one individual organism's lifetime; however, these epigenetic changes can be transmitted to the organism's offspring through a process called
transgenerational epigenetic inheritance
Transgenerational epigenetic inheritance is the transmission of epigenetic markers from one organism to the next (i.e., from parent to child) that affects the traits of offspring without altering the nucleic acid primary structure, primary structur ...
. Moreover, if gene inactivation occurs in a sperm or egg cell that results in fertilization, this epigenetic modification may also be transferred to the next generation.
Specific epigenetic processes include
paramutation
In epigenetics, a paramutation is an interaction between two alleles at a single locus, whereby one allele induces a heritable change in the other allele. The change may be in the pattern of DNA methylation or histone modifications. The allele indu ...
,
bookmarking
Bookmarking (also "gene bookmarking" or "mitotic bookmarking") refers to a potential mechanism of transmission of gene expression programs through cell division.
During mitosis, gene transcription is silenced and most transcription factors are re ...
,
imprinting,
gene silencing,
X chromosome inactivation,
position effect
Position effect is the effect on the expression of a gene when its location in a chromosome is changed, often by translocation. This has been well described in ''Drosophila'' with respect to eye color and is known as position effect variegation ( ...
,
DNA methylation reprogramming
In biology, reprogramming refers to erasure and remodeling of epigenetic marks, such as DNA methylation, during mammalian development or in cell culture. Such control is also often associated with alternative covalent modifications of histones.
...
,
transvection,
maternal effects, the progress of
carcinogenesis, many effects of
teratogens, regulation of
histone modifications and
heterochromatin
Heterochromatin is a tightly packed form of DNA or '' condensed DNA'', which comes in multiple varieties. These varieties lie on a continue between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a role ...
, and technical limitations affecting
parthenogenesis and
cloning.
DNA damage
DNA damage can also cause epigenetic changes. DNA damage is very frequent, occurring on average about 60,000 times a day per cell of the human body (see
DNA damage (naturally occurring)). These damages are largely repaired, however, epigenetic changes can still remain at the site of DNA repair. In particular, a double strand break in DNA can initiate unprogrammed epigenetic gene silencing both by causing DNA methylation as well as by promoting silencing types of histone modifications (chromatin remodeling - see next section). In addition, the enzyme
Parp1 (poly(ADP)-ribose polymerase) and its product poly(ADP)-ribose (PAR) accumulate at sites of DNA damage as part of the repair process. This accumulation, in turn, directs recruitment and activation of the chromatin remodeling protein, ALC1, that can cause
nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone proteins and resembles thread wrapped around a spool. The nucleosome is the fundamen ...
remodeling. Nucleosome remodeling has been found to cause, for instance, epigenetic silencing of DNA repair gene MLH1.
DNA damaging chemicals, such as
benzene,
hydroquinone,
styrene,
carbon tetrachloride
Carbon tetrachloride, also known by many other names (such as tetrachloromethane, also IUPAC nomenclature of inorganic chemistry, recognised by the IUPAC, carbon tet in the cleaning industry, Halon-104 in firefighting, and Refrigerant-10 in HVAC ...
and
trichloroethylene, cause considerable hypomethylation of DNA, some through the activation of oxidative stress pathways.
Foods are known to alter the epigenetics of rats on different diets. Some food components epigenetically increase the levels of DNA repair enzymes such as
MGMT
MGMT () is an American indie rock band formed in 2002 in Middletown, Connecticut. It was founded by multi-instrumentalists Andrew VanWyngarden and Ben Goldwasser. Alongside VanWyngarden and Goldwasser, MGMT's live lineup currently consists of ...
and
MLH1
DNA mismatch repair protein Mlh1 or MutL protein homolog 1 is a protein that in humans is encoded by the MLH1 gene located on chromosome 3. It is a gene commonly associated with hereditary nonpolyposis colorectal cancer. Orthologs of human MLH1 h ...
and
p53. Other food components can reduce DNA damage, such as soy
isoflavones Isoflavones are substituted derivatives of isoflavone, a type of naturally occurring isoflavonoids, many of which act as phytoestrogens in mammals. Isoflavones are produced almost exclusively by the members of the bean family, Fabaceae (Leguminosae) ...
. In one study, markers for oxidative stress, such as modified nucleotides that can result from DNA damage, were decreased by a 3-week diet supplemented with soy. A decrease in oxidative DNA damage was also observed 2 h after consumption of
anthocyanin
Anthocyanins (), also called anthocyans, are water-soluble vacuolar pigments that, depending on their pH, may appear red, purple, blue, or black. In 1835, the German pharmacist Ludwig Clamor Marquart gave the name Anthokyan to a chemical compo ...
-rich
bilberry (''
Vaccinium myrtillius'' L.)
pomace extract.
DNA repair
Damage to DNA is very common and is constantly being repaired. Epigenetic alterations can accompany DNA repair of oxidative damage or double-strand breaks. In human cells, oxidative DNA damage occurs about 10,000 times a day and DNA double-strand breaks occur about 10 to 50 times a cell cycle in somatic replicating cells (see
DNA damage (naturally occurring)). The selective advantage of DNA repair is to allow the cell to survive in the face of DNA damage. The selective advantage of epigenetic alterations that occur with DNA repair is not clear.
Repair of oxidative DNA damage can alter epigenetic markers
In the steady state (with endogenous damages occurring and being repaired), there are about 2,400 oxidatively damaged guanines that form
8-oxo-2'-deoxyguanosine (8-OHdG) in the average mammalian cell DNA.
8-OHdG constitutes about 5% of the oxidative damages commonly present in DNA.
The oxidized guanines do not occur randomly among all guanines in DNA. There is a sequence preference for the guanine at a
methylated CpG site (a cytosine followed by guanine along its
5' → 3' direction and where the cytosine is methylated (5-mCpG)).
A 5-mCpG site has the lowest ionization potential for guanine oxidation.

Oxidized guanine has mispairing potential and is mutagenic.
Oxoguanine glycosylase
8-Oxoguanine glycosylase, also known as OGG1, is a DNA glycosylase enzyme that, in humans, is encoded by the ''OGG1'' gene. It is involved in base excision repair. It is found in bacterial, archaeal and eukaryotic species.
Function
OGG1 is th ...
(OGG1) is the primary enzyme responsible for the excision of the oxidized guanine during DNA repair. OGG1 finds and binds to an 8-OHdG within a few seconds.
However, OGG1 does not immediately excise 8-OHdG. In HeLa cells half maximum removal of 8-OHdG occurs in 30 minutes,
and in irradiated mice, the 8-OHdGs induced in the mouse liver are removed with a half-life of 11 minutes.
When OGG1 is present at an oxidized guanine within a methylated
CpG site it recruits
TET1 to the 8-OHdG lesion (see Figure). This allows TET1 to demethylate an adjacent methylated cytosine. Demethylation of cytosine is an epigenetic alteration.
As an example, when human mammary epithelial cells were treated with H
2O
2 for six hours, 8-OHdG increased about 3.5-fold in DNA and this caused about 80% demethylation of the 5-methylcytosines in the genome.
Demethylation of CpGs in a gene promoter by
TET enzyme activity increases transcription of the gene into messenger RNA.
In cells treated with H
2O
2, one particular gene was examined,
''BACE1''.
The methylation level of the ''BACE1''
CpG island
The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG isl ...
was reduced (an epigenetic alteration) and this allowed about 6.5 fold increase of expression of ''BACE1'' messenger RNA.
While six-hour incubation with H
2O
2 causes considerable demethylation of 5-mCpG sites, shorter times of H
2O
2 incubation appear to promote other epigenetic alterations. Treatment of cells with H
2O
2 for 30 minutes causes the mismatch repair protein heterodimer MSH2-MSH6 to recruit DNA methyltransferase 1 (DNMT1) to sites of some kinds of oxidative DNA damage.
This could cause increased methylation of cytosines (epigenetic alterations) at these locations.
Jiang et al.
treated
HEK 293 cells with agents causing oxidative DNA damage, (
potassium bromate
Potassium bromate (KBrO3), is a bromate of potassium and takes the form of white crystals or powder. It is a strong oxidizing agent.
It is a toxic and carcinogenic compound.
Preparation
Potassium bromate is produced when bromine is passed throug ...
(KBrO3) or
potassium chromate
Potassium chromate is the inorganic compound with the formula K2 CrO4. This yellow solid is the potassium salt of the chromate anion. It is a common laboratory chemical, whereas sodium chromate is important industrially.
Structure
Two crystal ...
(K2CrO4)).
Base excision repair (BER) of oxidative damage occurred with the DNA repair enzyme
polymerase beta localizing to oxidized guanines. Polymerase beta is the main human polymerase in short-patch BER of oxidative DNA damage. Jiang et al.
also found that polymerase beta recruited the
DNA methyltransferase protein DNMT3b to BER repair sites. They then evaluated the methylation pattern at the single nucleotide level in a small region of DNA including the
promoter region and the early transcription region of the
BRCA1 gene. Oxidative DNA damage from bromate modulated the DNA methylation pattern (caused epigenetic alterations) at CpG sites within the region of DNA studied. In untreated cells, CpGs located at −189, −134, −29, −19, +16, and +19 of the BRCA1 gene had methylated cytosines (where numbering is from the
messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the p ...
transcription start site, and negative numbers indicate nucleotides in the upstream
promoter region). Bromate treatment-induced oxidation resulted in the loss of cytosine methylation at −189, −134, +16 and +19 while also leading to the formation of new methylation at the CpGs located at −80, −55, −21 and +8 after DNA repair was allowed.
Homologous recombinational repair alters epigenetic markers
At least four articles report the recruitment of
DNA methyltransferase 1 (DNMT1) to sites of DNA double-strand breaks.
During
homologous recombinational repair (HR) of the double-strand break, the involvement of DNMT1 causes the two repaired strands of DNA to have different levels of methylated cytosines. One strand becomes frequently methylated at about 21
CpG sites downstream of the repaired double-strand break. The other DNA strand loses methylation at about six CpG sites that were previously methylated downstream of the double-strand break, as well as losing methylation at about five CpG sites that were previously methylated upstream of the double-strand break. When the chromosome is replicated, this gives rise to one daughter chromosome that is heavily methylated downstream of the previous break site and one that is unmethylated in the region both upstream and downstream of the previous break site. With respect to the gene that was broken by the double-strand break, half of the progeny cells express that gene at a high level and in the other half of the progeny cells expression of that gene is repressed. When clones of these cells were maintained for three years, the new methylation patterns were maintained over that time period.
In mice with a CRISPR-mediated homology-directed recombination insertion in their genome there were a large number of increased methylations of CpG sites within the double-strand break-associated insertion.
Non-homologous end joining can cause some epigenetic marker alterations
Non-homologous end joining (NHEJ) repair of a double-strand break can cause a small number of demethylations of pre-existing cytosine DNA methylations downstream of the repaired double-strand break.
Further work by Allen et al.
showed that NHEJ of a DNA double-strand break in a cell could give rise to some progeny cells having repressed expression of the gene harboring the initial double-strand break and some progeny having high expression of that gene due to epigenetic alterations associated with NHEJ repair. The frequency of epigenetic alterations causing repression of a gene after an NHEJ repair of a DNA double-strand break in that gene may be about 0.9%.
Techniques used to study epigenetics
Epigenetic research uses a wide range of
molecular biological
Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions. The study of chemical and physi ...
techniques to further understanding of epigenetic phenomena. These techniques include
chromatin immunoprecipitation Chromatin immunoprecipitation (ChIP) is a type of immunoprecipitation experimental technique used to investigate the interaction between proteins and DNA in the cell. It aims to determine whether specific proteins are associated with specific genom ...
(together with its large-scale variants
ChIP-on-chip and
ChIP-Seq),
fluorescent in situ hybridization
Fluorescence ''in situ'' hybridization (FISH) is a molecular cytogenetic technique that uses fluorescent probes that bind to only particular parts of a nucleic acid sequence with a high degree of sequence complementarity. It was developed ...
, methylation-sensitive
restriction enzymes, DNA adenine methyltransferase identification (
DamID) and
bisulfite sequencing
Bisulfite sequencing (also known as bisulphite sequencing) is the use of bisulfite treatment of DNA before routine sequencing to determine the pattern of methylation. DNA methylation was the first discovered epigenetic mark, and remains the mo ...
.
Furthermore, the use of
bioinformatics
Bioinformatics () is an interdisciplinary field that develops methods and software tools for understanding biological data, in particular when the data sets are large and complex. As an interdisciplinary field of science, bioinformatics combi ...
methods has a role in
computational epigenetics
Computational epigenetics
uses statistical methods and mathematical modelling in epigenetic research. Due to the recent explosion of epigenome datasets, computational methods play an increasing role in all areas of epigenetic research.
Definition ...
.
[
]
Chromatin Immunoprecipitation
Chromatin Immunoprecipitation (ChIP) has helped bridge the gap between DNA and epigenetic interactions.
Fluorescent ''in situ'' hybridization
Fluorescent ''in situ'' hybridization (FISH) is very important to understand epigenetic mechanisms.
Methylation-sensitive restriction enzymes
Methylation sensitive restriction enzymes paired with PCR is a way to evaluate methylation in DNA - specifically the CpG sites.
Bisulfite sequencing
Bisulfite sequencing is another way to evaluate DNA methylation. Cytosine will be changed to uracil from being treated with sodium bisulfite, whereas methylated cytosines will not be affected.
Mechanisms
Epigenetic mechanisms are sensitive to environmental effects, and key participants in shaping an adult phenotype. Several types of epigenetic inheritance systems may play a role in what has become known as cell memory,
Covalent modifications
Covalent modification of either DNA (e.g. cytosine methylation and hydroxymethylation) or of histone proteins (e.g. lysine acetylation, lysine and arginine methylation, serine and threonine phosphorylation, and lysine ubiquitination and sumoylation) play central roles in many types of epigenetic inheritance. Therefore, the word "epigenetics" is sometimes used as a synonym for these processes. However, this can be misleading. Chromatin remodeling is not always inherited, and not all epigenetic inheritance involves chromatin remodeling. Because the phenotype of a cell or individual is affected by which of its genes are transcribed, heritable transcription states can give rise to epigenetic effects. There are several layers of regulation of
Because the phenotype of a cell or individual is affected by which of its genes are transcribed, heritable transcription states can give rise to epigenetic effects. There are several layers of regulation of gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, protein or non-coding RNA, and ultimately affect a phenotype, as the final effect. The ...
. One way that genes are regulated is through the remodeling of chromatin. Chromatin is the complex of DNA and the histone proteins with which it associates. If the way that DNA is wrapped around the histones changes, gene expression can change as well. Chromatin remodeling is accomplished through two main mechanisms:
# The first way is post translational modification of the amino acids that make up histone proteins. Histone proteins are made up of long chains of amino acids. If the amino acids that are in the chain are changed, the shape of the histone might be modified. DNA is not completely unwound during replication. It is possible, then, that the modified histones may be carried into each new copy of the DNA. Once there, these histones may act as templates, initiating the surrounding new histones to be shaped in the new manner. By altering the shape of the histones around them, these modified histones would ensure that a lineage-specific transcription program is maintained after cell division.
# The second way is the addition of methyl groups to the DNA, mostly at CpG sites, to convert cytosine to 5-methylcytosine. 5-Methylcytosine performs much like a regular cytosine, pairing with a guanine in double-stranded DNA. However, when methylated cytosines are present in CpG sites in the promoter and enhancer regions of genes, the genes are often repressed.coding region
The coding region of a gene, also known as the coding sequence (CDS), is the portion of a gene's DNA or RNA that codes for protein. Studying the length, composition, regulation, splicing, structures, and functions of coding regions compared to no ...
excluding the transcription start site) expression of the gene is often enhanced. Transcription of a gene usually depends on a transcription factor binding to a (10 base or less) recognition sequence at the enhancer that interacts with the promoter region of that gene ( Gene expression # Enhancers, transcription factors, mediator complex and DNA loops in mammalian transcription).methyl-CpG-binding domain
The Methyl-CpG-binding domain (MBD) in molecular biology binds to DNA that contains one or more symmetrically methylated CpGs. MBD has negligible non-specific affinity for unmethylated DNA. In vitro foot-printing with the chromosomal protein MeCP ...
(MBD) proteins. All MBDs interact with nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone proteins and resembles thread wrapped around a spool. The nucleosome is the fundamen ...
remodeling and histone deacetylase
Histone deacetylases (, HDAC) are a class of enzymes that remove acetyl groups (O=C-CH3) from an ε-N-acetyl lysine amino acid on a histone, allowing the histones to wrap the DNA more tightly. This is important because DNA is wrapped around his ...
complexes, which leads to gene silencing. In addition, another covalent modification involving methylated cytosine is its demethylation by TET enzymes. Hundreds of such demethylations occur, for instance, during learning and memory forming events in neurons.UHRF1
Ubiquitin-like, containing PHD and RING finger domains, 1, also known as UHRF1, is a protein which in humans is encoded by the ''UHRF1'' gene.
Function
This gene encodes a member of a subfamily of RING finger domain, RING-finger type E3 ubiqui ...
. UHRF1 has been recently recognized as essential for DNMT1-mediated maintenance of DNA methylation. UHRF1 is the protein that specifically recognizes hemi-methylated DNA, therefore bringing DNMT1 to its substrate to maintain DNA methylation. Although histone modifications occur throughout the entire sequence, the unstructured N-termini of histones (called histone tails) are particularly highly modified. These modifications include
Although histone modifications occur throughout the entire sequence, the unstructured N-termini of histones (called histone tails) are particularly highly modified. These modifications include acetylation
:
In organic chemistry, acetylation is an organic esterification reaction with acetic acid. It introduces an acetyl group into a chemical compound. Such compounds are termed ''acetate esters'' or simply '' acetates''. Deacetylation is the oppo ...
, methylation
In the chemical sciences, methylation denotes the addition of a methyl group on a substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These t ...
, ubiquitylation, phosphorylation
In chemistry, phosphorylation is the attachment of a phosphate group to a molecule or an ion. This process and its inverse, dephosphorylation, are common in biology and could be driven by natural selection. Text was copied from this source, wh ...
, sumoylation
In molecular biology, SUMO (Small Ubiquitin-like Modifier) proteins are a family of small proteins that are covalently attached to and detached from other proteins in cells to modify their function. This process is called SUMOylation (sometimes w ...
, ribosylation and citrullination. Acetylation is the most highly studied of these modifications. For example, acetylation of the K14 and K9 lysine
Lysine (symbol Lys or K) is an α-amino acid that is a precursor to many proteins. It contains an α-amino group (which is in the protonated form under biological conditions), an α-carboxylic acid group (which is in the deprotonated −C ...
s of the tail of histone H3 by histone acetyltransferase enzymes (HATs) is generally related to transcriptional competence (see Figure).
One mode of thinking is that this tendency of acetylation to be associated with "active" transcription is biophysical in nature. Because it normally has a positively charged nitrogen at its end, lysine can bind the negatively charged phosphates of the DNA backbone. The acetylation event converts the positively charged amine group on the side chain into a neutral amide linkage. This removes the positive charge, thus loosening the DNA from the histone. When this occurs, complexes like SWI/SNF and other transcriptional factors can bind to the DNA and allow transcription to occur. This is the "cis" model of the epigenetic function. In other words, changes to the histone tails have a direct effect on the DNA itself.
Another model of epigenetic function is the "trans" model. In this model, changes to the histone tails act indirectly on the DNA. For example, lysine acetylation may create a binding site for chromatin-modifying enzymes (or transcription machinery as well). This chromatin remodeler can then cause changes to the state of the chromatin. Indeed, a bromodomain – a protein domain that specifically binds acetyl-lysine – is found in many enzymes that help activate transcription, including the SWI/SNF complex. It may be that acetylation acts in this and the previous way to aid in transcriptional activation.
The idea that modifications act as docking modules for related factors is borne out by histone methylation
Histone methylation is a process by which methyl groups are transferred to amino acids of histone proteins that make up nucleosomes, which the DNA double helix wraps around to form chromosomes. Methylation of histones can either increase or decrea ...
as well. Methylation of lysine 9 of histone H3 has long been associated with constitutively transcriptionally silent chromatin (constitutive heterochromatin
Heterochromatin is a tightly packed form of DNA or '' condensed DNA'', which comes in multiple varieties. These varieties lie on a continue between the two extremes of constitutive heterochromatin and facultative heterochromatin. Both play a role ...
) (see bottom Figure). It has been determined that a chromodomain (a domain that specifically binds methyl-lysine) in the transcriptionally repressive protein HP1 recruits HP1 to K9 methylated regions. One example that seems to refute this biophysical model for methylation is that tri-methylation of histone H3 at lysine 4 is strongly associated with (and required for full) transcriptional activation (see top Figure). Tri-methylation, in this case, would introduce a fixed positive charge on the tail.
It has been shown that the histone lysine methyltransferase (KMT) is responsible for this methylation activity in the pattern of histones H3 & H4. This enzyme utilizes a catalytically active site called the SET domain (Suppressor of variegation, Enhancer of Zeste, Trithorax). The SET domain is a 130-amino acid sequence involved in modulating gene activities. This domain has been demonstrated to bind to the histone tail and causes the methylation of the histone.nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone proteins and resembles thread wrapped around a spool. The nucleosome is the fundamen ...
. The idea that multiple dynamic modifications regulate gene transcription in a systematic and reproducible way is called the histone code
The histone code is a hypothesis that the transcription of genetic information encoded in DNA is in part regulated by chemical modifications (known as ''histone marks'') to histone proteins, primarily on their unstructured ends. Together with sim ...
, although the idea that histone state can be read linearly as a digital information carrier has been largely debunked. One of the best-understood systems that orchestrate chromatin-based silencing is the SIR protein Silent Information Regulator (SIR) proteins are involved in regulating gene expression. SIR proteins organize heterochromatin near telomeres, rDNA, and at silent loci including hidden mating type loci in yeast. The SIR family of genes encodes cat ...
based silencing of the yeast hidden mating-type loci HML and HMR.
DNA methylation frequently occurs in repeated sequences, and helps to suppress the expression and mobility of ' transposable elements':CpG islands
The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG isl ...
where they remain unmethylated. Epigenetic changes of this type thus have the potential to direct increased frequencies of permanent genetic mutation. DNA methylation
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts t ...
patterns are known to be established and modified in response to environmental factors by a complex interplay of at least three independent DNA methyltransferases, DNMT1, DNMT3A, and DNMT3B, the loss of any of which is lethal in mice.proliferating cell nuclear antigen
Proliferating cell nuclear antigen (PCNA) is a DNA clamp that acts as a processivity factor for DNA polymerase δ in eukaryotic cells and is essential for replication. PCNA is a homotrimer and achieves its processivity by encircling the DNA, whe ...
(PCNA).covalent modification
Post-translational modification (PTM) is the covalent and generally enzymatic modification of proteins following protein biosynthesis. This process occurs in the endoplasmic reticulum and the golgi apparatus. Proteins are synthesized by ribosomes ...
s of histones.nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone proteins and resembles thread wrapped around a spool. The nucleosome is the fundamen ...
s recruit enzymes that similarly modify nearby nucleosomes. A simplified stochastic model for this type of epigenetics is found here.
RNA transcripts
Sometimes a gene, after being turned on, transcribes a product that (directly or indirectly) maintains the activity of that gene. For example, Hnf4
HNF4 (Hepatocyte Nuclear Factor 4) is a nuclear receptor protein mostly expressed in the liver, gut, kidney, and pancreatic beta cells that is critical for liver development. In humans, there are two isoforms of HNF4, HNF4α and HNF4γ, encod ...
and MyoD enhance the transcription of many liver-specific and muscle-specific genes, respectively, including their own, through the transcription factor activity of the proteins
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, respo ...
they encode. RNA signalling includes differential recruitment of a hierarchy of generic chromatin modifying complexes and DNA methyltransferases to specific loci by RNAs during differentiation and development.RNA
Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and deoxyribonucleic acid ( DNA) are nucleic acids. Along with lipids, proteins, and carbohydra ...
, or by formation of double-stranded RNA ( RNAi). Descendants of the cell in which the gene was turned on will inherit this activity, even if the original stimulus for gene-activation is no longer present. These genes are often turned on or off by signal transduction
Signal transduction is the process by which a chemical or physical signal is transmitted through a cell as a series of molecular events, most commonly protein phosphorylation catalyzed by protein kinases, which ultimately results in a cellula ...
, although in some systems where syncytia or gap junction
Gap junctions are specialized intercellular connections between a multitude of animal cell-types. They directly connect the cytoplasm of two cells, which allows various molecules, ions and electrical impulses to directly pass through a regulate ...
s are important, RNA may spread directly to other cells or nuclei by diffusion. A large amount of RNA and protein is contributed to the zygote by the mother during oogenesis
Oogenesis, ovogenesis, or oögenesis is the differentiation of the ovum (egg cell) into a cell competent to further develop when fertilized. It is developed from the primary oocyte by maturation. Oogenesis is initiated in the embryonic stage.
O ...
or via nurse cells, resulting in maternal effect phenotypes. A smaller quantity of sperm RNA is transmitted from the father, but there is recent evidence that this epigenetic information can lead to visible changes in several generations of offspring.
MicroRNAs
MicroRNAs (miRNAs) are members of non-coding RNAs that range in size from 17 to 25 nucleotides. miRNAs regulate a large variety of biological functions in plants and animals.CpG island
The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG isl ...
s,
mRNA
In 2011, it was demonstrated that the methylation
In the chemical sciences, methylation denotes the addition of a methyl group on a substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These t ...
of mRNA plays a critical role in human energy homeostasis. The obesity-associated FTO gene is shown to be able to demethylate Demethylating agents are chemical substances that can inhibit methylation, resulting in the expression of the previously hypermethylated silenced genes (see Methylation#Cancer for more detail). Cytidine analogs such as 5-azacytidine (azacitidine) ...
N6-methyladenosine in RNA.
sRNAs
sRNAs are small (50–250 nucleotides), highly structured, non-coding RNA fragments found in bacteria. They control gene expression including virulence genes in pathogens and are viewed as new targets in the fight against drug-resistant bacteria. They play an important role in many biological processes, binding to mRNA and protein targets in prokaryotes. Their phylogenetic analyses, for example through sRNA–mRNA target interactions or protein binding properties, are used to build comprehensive databases. sRNA- gene maps based on their targets in microbial genomes are also constructed.
Prions
Prion
Prions are misfolded proteins that have the ability to transmit their misfolded shape onto normal variants of the same protein. They characterize several fatal and transmissible neurodegenerative diseases in humans and many other animals. It ...
s are infectious forms of proteins. In general, proteins fold into discrete units that perform distinct cellular functions, but some proteins are also capable of forming an infectious conformational state known as a prion. Although often viewed in the context of infectious disease, prions are more loosely defined by their ability to catalytically convert other native state versions of the same protein to an infectious conformational state. It is in this latter sense that they can be viewed as epigenetic agents capable of inducing a phenotypic change without a modification of the genome.
Fungal prion
A fungal prion is a prion that infects fungal hosts. Fungal prions are naturally occurring proteins that can switch between multiple, structurally distinct conformations, at least one of which is self-propagating and transmissible to other prion ...
s are considered by some to be epigenetic because the infectious phenotype caused by the prion can be inherited without modification of the genome. PSI+ and URE3, discovered in yeast in 1965 and 1971, are the two best studied of this type of prion.codon
The genetic code is the set of rules used by living cells to translate information encoded within genetic material ( DNA or RNA sequences of nucleotide triplets, or codons) into proteins. Translation is accomplished by the ribosome, which links ...
s, an effect that results in suppression of nonsense mutations in other genes.
Structural inheritance
In ciliates such as '' Tetrahymena'' and '' Paramecium'', genetically identical cells show heritable differences in the patterns of ciliary rows on their cell surface. Experimentally altered patterns can be transmitted to daughter cells. It seems existing structures act as templates for new structures. The mechanisms of such inheritance are unclear, but reasons exist to assume that multicellular organisms also use existing cell structures to assemble new ones.
Nucleosome positioning
Eukaryotic genomes have numerous nucleosomes. Nucleosome position is not random, and determine the accessibility of DNA to regulatory proteins. Promoters active in different tissues have been shown to have different nucleosome positioning features. This determines differences in gene expression and cell differentiation. It has been shown that at least some nucleosomes are retained in sperm cells (where most but not all histones are replaced by protamines). Thus nucleosome positioning is to some degree inheritable. Recent studies have uncovered connections between nucleosome positioning and other epigenetic factors, such as DNA methylation and hydroxymethylation.
Histone variants
Different histone variants
Histone variants are proteins that substitute for the core canonical histones ( H3, H4, H2A, H2B) in nucleosomes in eukaryotes and often confer specific structural and functional features. The term might also include a set of linker histone (H1 ...
are incorporated into specific regions of the genome non-randomly. Their differential biochemical characteristics can affect genome functions via their roles in gene regulation, and maintenance of chromosome structures.
Genomic architecture
The three-dimensional configuration of the genome (the 3D genome) is complex, dynamic and crucial for regulating genomic function and nuclear processes such as DNA replication, transcription and DNA-damage repair.
Functions and consequences
In the brain
Memory
Memory formation
Memory is the faculty of the mind by which data or information is encoded, stored, and retrieved when needed. It is the retention of information over time for the purpose of influencing future action. If past events could not be remembered, ...
and maintenance are due to epigenetic alterations that cause the required dynamic changes in gene transcription that create and renew memory in neurons.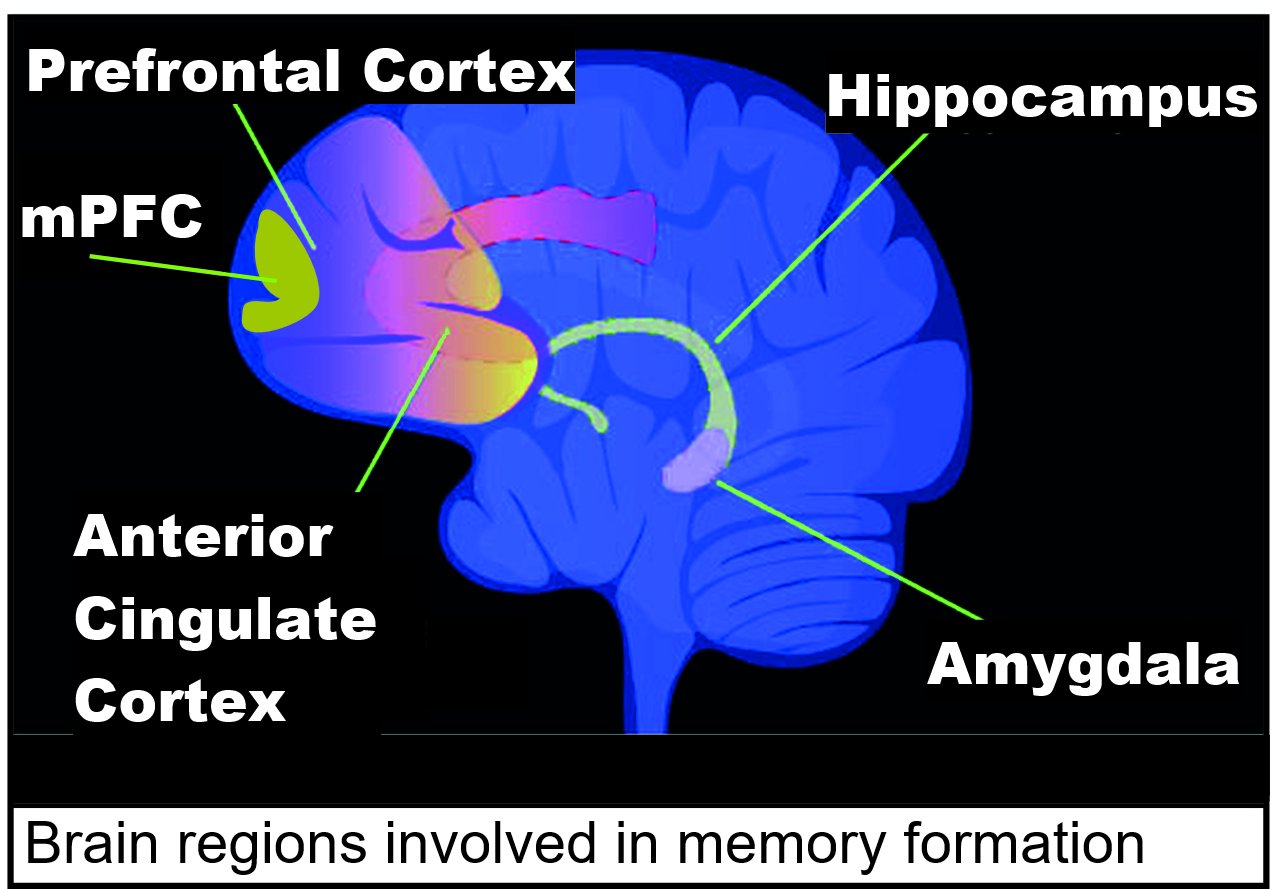 Areas of the brain important in the formation of memories include the hippocampus, medial prefrontal cortex (mPFC), anterior cingulate cortex and amygdala, as shown in the diagram of the human brain in this section.
Areas of the brain important in the formation of memories include the hippocampus, medial prefrontal cortex (mPFC), anterior cingulate cortex and amygdala, as shown in the diagram of the human brain in this section.immediate early genes
Immediate early genes (IEGs) are genes which are activated transiently and rapidly in response to a wide variety of cellular stimuli. They represent a standing response mechanism that is activated at the transcription level in the first round of ...
(IEGs) that are important in memory formation, allowing their expression in mRNA, with peak mRNA transcription at seven to ten minutes after CFC.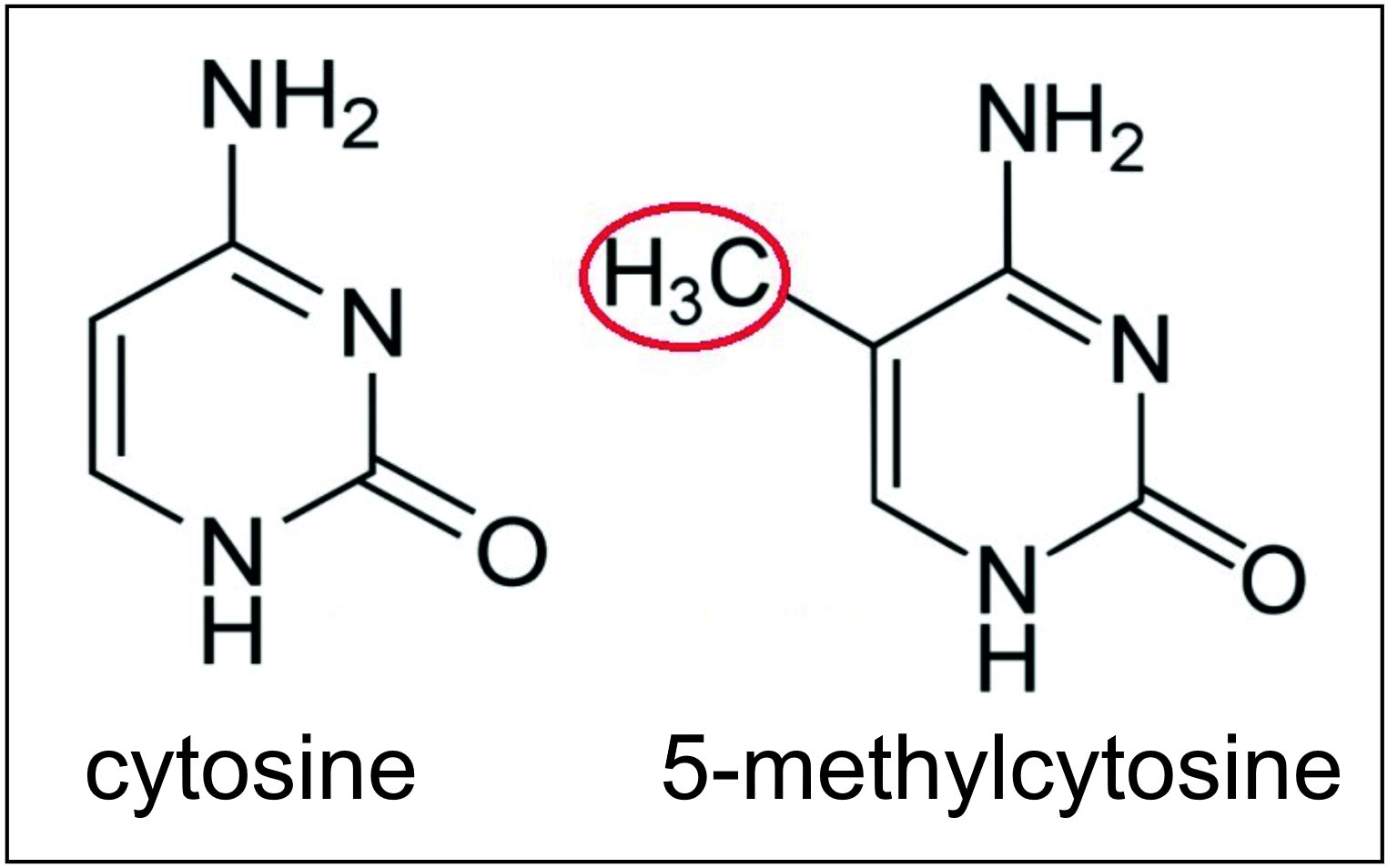 The DNA methyltransferases DNMT3A1, DNMT3A2 and DNMT3B can all methylate cytosines (see image this section) at CpG sites in or near the promoters of genes. As shown by Manzo et al.,
The DNA methyltransferases DNMT3A1, DNMT3A2 and DNMT3B can all methylate cytosines (see image this section) at CpG sites in or near the promoters of genes. As shown by Manzo et al., mRNAs
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the p ...
are often packaged into neuronal granules, or messenger RNP
Messenger RNP (messenger ribonucleoprotein) is mRNA with bound proteins. mRNA does not exist "naked" ''in vivo'' but is always bound by various proteins while being synthesized, spliced, exported, and translated in the cytoplasm.
Messenger RNPs we ...
, consisting of mRNA, small and large ribosomal subunits, translation initiation factors and RNA-binding proteins that regulate mRNA function. These neuronal granules are transported from the neuron nucleus and are directed, according to 3′ untranslated regions of the mRNA in the granules (their "zip codes"), to neuronal dendrites. Roughly 2,500 mRNAs may be localized to the dendrites of hippocampal pyramidal neurons and perhaps 450 transcripts are in excitatory presynaptic nerve terminals (dendritic spines). The altered assortments of transcripts (dependent on epigenetic alterations in the neuron nucleus) have different sensitivities in response to signals, which is the basis of altered synaptic plasticity. Altered synaptic plasticity is often considered the neurochemical foundation of learning and memory.
Other and general
In adulthood, changes in the epigenome are important for various higher cognitive functions. Dysregulation of epigenetic mechanisms is implicated in neurodegenerative disorders and diseases. Epigenetic modifications in neurons are dynamic and reversible. Epigenetic regulation impacts neuronal action, affecting learning, memory, and other cognitive processes.
Early events, including during embryonic development
An embryo is an initial stage of development of a multicellular organism. In organisms that reproduce sexually, embryonic development is the part of the life cycle that begins just after fertilization of the female egg cell by the male sperm ...
, can influence development, cognition, and health outcomes through epigenetic mechanisms.
Epigenetic mechanisms have been proposed as "a potential molecular mechanism for effects of endogenous hormones on the organization of developing brain circuits".
Nutrients could interact with the epigenome to "protect or boost cognitive processes across the lifespan".
Epigenetics play a major role in brain aging
Aging is a major risk factor for most common neurodegenerative diseases, including mild cognitive impairment, dementias including Alzheimer's disease, cerebrovascular disease, Parkinson's disease, and Lou Gehrig's disease. While much research has ...
and age-related cognitive decline, with relevance to life extension
Life extension is the concept of extending the human life expectancy, lifespan, either modestly through improvements in medicine or dramatically by increasing the maximum lifespan beyond its generally-settled oldest people, limit of 125 years.
S ...
.
A review suggests neurobiological effects of physical exercise via epigenetics seem "central to building an 'epigenetic memory' to influence long-term brain function and behavior" and may even be heritable.
With the axo-ciliary synapse
In the nervous system, a synapse is a structure that permits a neuron (or nerve cell) to pass an electrical or chemical signal to another neuron or to the target effector cell.
Synapses are essential to the transmission of nervous impulses from ...
, there is communication between serotonergic axons and antenna-like primary cilia
The cilium, plural cilia (), is a membrane-bound organelle found on most types of eukaryotic cell, and certain microorganisms known as ciliates. Cilia are absent in bacteria and archaea. The cilium has the shape of a slender threadlike projecti ...
of CA1 pyramidal
A pyramid (from el, πυραμίς ') is a structure whose outer surfaces are triangular and converge to a single step at the top, making the shape roughly a pyramid in the geometric sense. The base of a pyramid can be trilateral, quadrilater ...
neurons that alters the neuron's epigenetic
In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the DNA sequence. The Greek prefix '' epi-'' ( "over, outside of, around") in ''epigenetics'' implies features that are "o ...
state in the nucleus
Nucleus ( : nuclei) is a Latin word for the seed inside a fruit. It most often refers to:
*Atomic nucleus, the very dense central region of an atom
*Cell nucleus, a central organelle of a eukaryotic cell, containing most of the cell's DNA
Nucle ...
via the signalling distinct from that at the plasma membrane
The cell membrane (also known as the plasma membrane (PM) or cytoplasmic membrane, and historically referred to as the plasmalemma) is a biological membrane that separates and protects the interior of all cells from the outside environment (t ...
(and longer-term).
Epigenetics also play a major role in the brain evolution in and to humans.
Development
Developmental epigenetics can be divided into predetermined and probabilistic epigenesis. Predetermined epigenesis is a unidirectional movement from structural development in DNA to the functional maturation of the protein. "Predetermined" here means that development is scripted and predictable. Probabilistic epigenesis on the other hand is a bidirectional structure-function development with experiences and external molding development.nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone proteins and resembles thread wrapped around a spool. The nucleosome is the fundamen ...
repositioning, is very important in the development of multicellular eukaryotic organisms.mammal
Mammals () are a group of vertebrate animals constituting the class Mammalia (), characterized by the presence of mammary glands which in females produce milk for feeding (nursing) their young, a neocortex (a region of the brain), fur or ...
s, some stem cells continue producing newly differentiated cells throughout life, such as in neurogenesis
Neurogenesis is the process by which nervous system cells, the neurons, are produced by neural stem cells (NSCs). It occurs in all species of animals except the porifera (sponges) and placozoans. Types of NSCs include neuroepithelial cells (NECs) ...
, but mammals are not able to respond to loss of some tissues, for example, the inability to regenerate limbs, which some other animals are capable of. Epigenetic modifications regulate the transition from neural stem cells to glial progenitor cells (for example, differentiation into oligodendrocytes is regulated by the deacetylation and methylation of histones. Unlike animals, plant cells do not terminally differentiate, remaining totipotent with the ability to give rise to a new individual plant. While plants do utilize many of the same epigenetic mechanisms as animals, such as chromatin remodeling
Chromatin remodeling is the dynamic modification of chromatin architecture to allow access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby control gene expression. Such remodeling is principally carried out ...
, it has been hypothesized that some kinds of plant cells do not use or require "cellular memories", resetting their gene expression patterns using positional information from the environment and surrounding cells to determine their fate.diabetic embryopathy
Diabetic embryopathy refers to congenital maldevelopments that are linked to maternal diabetes. Prenatal exposure to hyperglycemia can result in spontaneous abortions, perinatal mortality, and malformations. Type 1 and Type 2 diabetic pregnanci ...
, on methylation
In the chemical sciences, methylation denotes the addition of a methyl group on a substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These t ...
signatures.
Controversial results from one study suggested that traumatic experiences might produce an epigenetic signal that is capable of being passed to future generations. Mice were trained, using foot shocks, to fear a cherry blossom odor. The investigators reported that the mouse offspring had an increased aversion to this specific odor. They suggested epigenetic changes that increase gene expression, rather than in DNA itself, in a gene, M71, that governs the functioning of an odor receptor in the nose that responds specifically to this cherry blossom smell. There were physical changes that correlated with olfactory (smell) function in the brains of the trained mice and their descendants. Several criticisms were reported, including the study's low statistical power as evidence of some irregularity such as bias in reporting results.
Transgenerational
Epigenetic mechanisms were a necessary part of the evolutionary origin of cell differentiation
Cellular differentiation is the process in which a stem cell alters from one type to a differentiated one. Usually, the cell changes to a more specialized type. Differentiation happens multiple times during the development of a multicellula ...
.paramutation
In epigenetics, a paramutation is an interaction between two alleles at a single locus, whereby one allele induces a heritable change in the other allele. The change may be in the pattern of DNA methylation or histone modifications. The allele indu ...
observed in maize). Although most of these multigenerational epigenetic traits are gradually lost over several generations, the possibility remains that multigenerational epigenetics could be another aspect to evolution and adaptation.
As mentioned above, some define epigenetics as heritable.
A sequestered germ line or Weismann barrier is specific to animals, and epigenetic inheritance is more common in plants and microbes. Eva Jablonka
Eva Jablonka ( he, חווה יבלונקה) (born 1952) is an Israeli evolutionary theorist and geneticist, known especially for her interest in epigenetic inheritance. Born in 1952 in Poland, she emigrated to Israel in 1957. She is a professor a ...
, Marion J. Lamb
Marion Julia Lamb (29 July 1939 – 12 December 2021) was Senior Lecturer at Birkbeck, University of London, before her retirement. She studied the effect of environmental conditions such as heat, radiation and pollution on metabolic activity an ...
and Étienne Danchin have argued that these effects may require enhancements to the standard conceptual framework of the modern synthesis and have called for an extended evolutionary synthesis.Thomas Dickins
Thomas may refer to:
People
* List of people with given name Thomas
* Thomas (name)
* Thomas (surname)
* Saint Thomas (disambiguation)
* Thomas Aquinas (1225–1274) Italian Dominican friar, philosopher, and Doctor of the Church
* Thomas the Ap ...
and Qazi Rahman state that epigenetic mechanisms such as DNA methylation and histone modification are genetically inherited under the control of natural selection and therefore fit under the earlier "modern synthesis".
Two important ways in which epigenetic inheritance can differ from traditional genetic inheritance, with important consequences for evolution, are:
* rates of epimutation can be much faster than rates of mutationtransgenerational epigenetic inheritance
Transgenerational epigenetic inheritance is the transmission of epigenetic markers from one organism to the next (i.e., from parent to child) that affects the traits of offspring without altering the nucleic acid primary structure, primary structur ...
phenomena have been reported in a wide range of organisms, including prokaryotes, plants, and animals.yeast prion
A fungal prion is a prion that infects fungal hosts. Fungal prions are naturally occurring proteins that can switch between multiple, structurally distinct conformations, at least one of which is self-propagating and transmissible to other prion ...
PSI is generated by a conformational change of a translation termination factor, which is then inherited by daughter cells. This can provide a survival advantage under adverse conditions, exemplifying epigenetic regulation which enables unicellular organisms to respond rapidly to environmental stress. Prions can be viewed as epigenetic agents capable of inducing a phenotypic change without modification of the genome.single molecule real time sequencing
Single-molecule real-time (SMRT) sequencing is a parallelized single molecule DNA sequencing method. Single-molecule real-time sequencing utilizes a zero-mode waveguide (ZMW). A single DNA polymerase enzyme is affixed at the bottom of a ZMW with a ...
, in which polymerase sensitivity allows for measuring methylation and other modifications as a DNA molecule is being sequenced. Several projects have demonstrated the ability to collect genome-wide epigenetic data in bacteria.
Epigenetics in bacteria
 While epigenetics is of fundamental importance in
While epigenetics is of fundamental importance in eukaryote
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacte ...
s, especially metazoans, it plays a different role in bacteria. Most importantly, eukaryotes use epigenetic mechanisms primarily to regulate gene expression which bacteria rarely do. However, bacteria make widespread use of postreplicative DNA methylation for the epigenetic control of DNA-protein interactions. Bacteria also use DNA adenine methylation (rather than DNA cytosine methylation) as an epigenetic signal. DNA adenine methylation is important in bacteria virulence in organisms such as '' Escherichia coli'', ''Salmonella
''Salmonella'' is a genus of rod-shaped (bacillus) Gram-negative bacteria of the family Enterobacteriaceae. The two species of ''Salmonella'' are ''Salmonella enterica'' and ''Salmonella bongori''. ''S. enterica'' is the type species and is fur ...
, Vibrio, Yersinia, Haemophilus'', and '' Brucella''. In '' Alphaproteobacteria'', methylation of adenine regulates the cell cycle and couples gene transcription to DNA replication. In ''Gammaproteobacteria
Gammaproteobacteria is a class of bacteria in the phylum Pseudomonadota (synonym Proteobacteria). It contains about 250 genera, which makes it the most genera-rich taxon of the Prokaryotes. Several medically, ecologically, and scientifically imp ...
'', adenine methylation provides signals for DNA replication, chromosome segregation, mismatch repair, packaging of bacteriophage, transposase activity and regulation of gene expression.
Medicine
Epigenetics has many and varied potential medical applications.Randy Jirtle
Randy Jirtle (born November 9, 1947) is an American biologist noted for his research in epigenetics, the branch of biology that deals with inherited information that does not reside in the nucleotide sequence of DNA. Jirtle retired from Duke Uni ...
, Ph.D., of Duke University Medical Center, think epigenetics may ultimately turn out to have a greater role in disease than genetics.
Twins
Direct comparisons of identical twins constitute an optimal model for interrogating environmental epigenetics. In the case of humans with different environmental exposures, monozygotic (identical) twins were epigenetically indistinguishable during their early years, while older twins had remarkable differences in the overall content and genomic distribution of 5-methylcytosine DNA and histone acetylation.Angelman syndrome
Angelman syndrome or Angelman's syndrome (AS) is a genetic disorder that mainly affects the nervous system. Symptoms include a small head and a specific facial appearance, severe intellectual disability, developmental disability, limited to no f ...
and Prader-Willi syndrome. These are normal genetic diseases caused by gene deletions or inactivation of the genes but are unusually common because individuals are essentially hemizygous because of genomic imprinting, and therefore a single gene knock out is sufficient to cause the disease, where most cases would require both copies to be knocked out.
Genomic imprinting
Some human disorders are associated with genomic imprinting, a phenomenon in mammals where the father and mother contribute different epigenetic patterns for specific genomic loci in their germ cells.Angelman syndrome
Angelman syndrome or Angelman's syndrome (AS) is a genetic disorder that mainly affects the nervous system. Symptoms include a small head and a specific facial appearance, severe intellectual disability, developmental disability, limited to no f ...
and Prader-Willi syndrome – both can be produced by the same genetic mutation, chromosome 15q partial deletion
Chromosome 15q partial deletion is a rare human genetic disorder, caused by a chromosomal aberration in which the long ("q") arm of one copy of chromosome 15 is deleted, or partially deleted. Like other chromosomal disorders, this increases the ri ...
, and the particular syndrome that will develop depends on whether the mutation is inherited from the child's mother or from their father.MeCP2
''MECP2'' (methyl CpG binding protein 2) is a gene that encodes the protein MECP2. MECP2 appears to be essential for the normal function of nerve cells. The protein seems to be particularly important for mature nerve cells, where it is present in ...
) is a transcriptional regulator that must be phosphorylated before releasing from the BDNF promoter, allowing transcription. Rett syndrome is underlain by mutations in the MeCP2 gene despite no large-scale changes in expression of MeCP2 being found in microarray analyses. BDNF is downregulated in the MECP2 mutant resulting in Rett syndrome, as well as the increase of early neural senescence
Senescence () or biological aging is the gradual deterioration of functional characteristics in living organisms. The word ''senescence'' can refer to either cellular senescence or to senescence of the whole organism. Organismal senescence inv ...
and accumulation of damaged DNA.[A person's paternal grandson is the son of a son of that person; a maternal grandson is the son of a daughter.] of Swedish men who were exposed during preadolescence to famine in the 19th century were less likely to die of cardiovascular disease. If food was plentiful, then diabetes mortality in the grandchildren increased, suggesting that this was a transgenerational epigenetic inheritance.[ Robert Winston refers to this study in a ] The opposite effect was observed for females – the paternal (but not maternal) granddaughters of women who experienced famine while in the womb (and therefore while their eggs were being formed) lived shorter lives on average.
Diabetic wound healing
Epigenetic modifications have given insight into the understanding of the pathophysiology of different disease conditions. Though, they are strongly associated with cancer, their role in other pathological conditions are of equal importance. It appears that the hyperglycaemic environment could imprint such changes at the genomic level, that macrophages are primed towards a pro-inflammatory state and could fail to exhibit any phenotypic alteration towards the pro-healing type. This phenomenon of altered Macrophage Polarization is mostly associated with all the diabetic complications in a clinical set-up. As of 2018, several reports reveal the relevance of different epigenetic modifications with respect to diabetic complications. Sooner or later, with the advancements in biomedical tools, the detection of such biomarkers as prognostic and diagnostic tools in patients could possibly emerge out as alternative approaches. It is noteworthy to mention here that the use of epigenetic modifications as therapeutic targets warrant extensive preclinical as well as clinical evaluation prior to use.
Examples of drugs altering gene expression from epigenetic events
The use of beta-lactam antibiotics can alter glutamate receptor activity and the action of cyclosporine on multiple transcription factors. Additionally, lithium can impact autophagy of aberrant proteins, and opioid drugs via chronic use can increase the expression of genes associated with addictive phenotypes.
In a groundbreaking 2003 report, Caspi and colleagues demonstrated that in a robust cohort of over one-thousand subjects assessed multiple times from preschool to adulthood, subjects who carried one or two copies of the short allele of the serotonin
Serotonin () or 5-hydroxytryptamine (5-HT) is a monoamine neurotransmitter. Its biological function is complex and multifaceted, modulating mood, cognition, reward, learning, memory, and numerous physiological processes such as vomiting and vas ...
transporter promoter polymorphism exhibited higher rates of adult depression and suicidality when exposed to childhood maltreatment when compared to long allele homozygotes with equal ELS exposure.
Addiction
Addiction is a disorder of the brain's reward system
The reward system (the mesocorticolimbic circuit) is a group of neural structures responsible for incentive salience (i.e., "wanting"; desire or craving for a reward and motivation), associative learning (primarily positive reinforcement and class ...
which arises through transcriptional
Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules calle ...
and neuroepigenetic mechanisms and occurs over time from chronically high levels of exposure to an addictive stimulus (e.g., morphine, cocaine, sexual intercourse, gambling, etc.).
Depression
Epigenetic inheritance of depression-related phenotypes has also been reported in a preclinical study.
Research
The two forms of heritable information, namely genetic and epigenetic, are collectively called dual inheritance. Members of the APOBEC/AID family of cytosine deaminase
In enzymology, a cytosine deaminase () is an enzyme that catalyzes the chemical reaction
:cytosine + H2O \rightleftharpoons uracil + NH3
Thus, the two substrates of this enzyme are cytosine and H2O, whereas its two products are uracil and NH3. ...
s may concurrently influence genetic and epigenetic inheritance using similar molecular mechanisms, and may be a point of crosstalk between these conceptually compartmentalized processes.
Epigenome editing
Epigenetic regulation of gene expression that could be altered or used in epigenome editing are or include mRNA/lncRNA modification, DNA methylation modification and histone modification
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn ar ...
.
CpG sites, SNPs and biological traits
Methylation is a widely characterized mechanism of genetic regulation that can determine biological traits. However, strong experimental evidences correlate methylation patterns in SNPs as an important additional feature for the classical activation/inhibition epigenetic dogma. Molecular interaction data, supported by colocalization analyses, identify multiple nuclear regulatory pathways, linking sequence variation to disturbances in DNA methylation and molecular and phenotypic variation.
''UBASH3B'' locus
''UBASH3B'' encodes a protein with tyrosine phosphatase activity, which has been previously linked to advanced neoplasia. SNP rs7115089 was identified as influencing DNA methylation and expression of this locus, as well as and Body Mass Index (BMI).
''NFKBIE'' locus
SNP rs730775 is located in the first intron of ''NFKBIE'' and is a ''cis'' eQTL for ''NFKBIE'' in whole blood.
''FADS1'' locus
Fatty acid desaturase 1 (FADS1) is a key enzyme in the metabolism of fatty acids. Moreover, rs174548 in the ''FADS1'' gene shows increased correlation with DNA methylation in people with high abundance of CD8+ T cells.
Pseudoscience
As epigenetics is in the early stages of development as a science and is surrounded by sensationalism in the public media, David Gorski
David Henry Gorski is an American surgical oncologist, professor of surgery at Wayne State University School of Medicine, and a surgical oncologist at the Barbara Ann Karmanos Cancer Institute, specializing in breast cancer surgery. He is an out ...
and geneticist Adam Rutherford have advised caution against the proliferation of false and pseudoscientific
Pseudoscience consists of statements, beliefs, or practices that claim to be both scientific and factual but are incompatible with the scientific method. Pseudoscience is often characterized by contradictory, exaggerated or unfalsifiable claim ...
conclusions by new age authors making unfounded suggestions that a person's genes and health can be manipulated by mind control. Misuse of the scientific term by quack authors has produced misinformation among the general public.
See also
* Baldwin effect
* Behavioral epigenetics
* Biological effects of radiation on the epigenome Ionizing radiation can cause biological effects which are passed on to offspring through the epigenome. The effects of radiation on cells has been found to be dependent on the dosage of the radiation, the location of the cell in regards to tissue ...
* Computational epigenetics
Computational epigenetics
uses statistical methods and mathematical modelling in epigenetic research. Due to the recent explosion of epigenome datasets, computational methods play an increasing role in all areas of epigenetic research.
Definition ...
* Contribution of epigenetic modifications to evolution Epigenetics is the study of changes in gene expression that occur via mechanisms such as DNA methylation, histone acetylation, and microRNA modification. When these epigenetic changes are Heritability, heritable, they can influence evolution. Curren ...
* DAnCER
Dance is a performing art form consisting of sequences of movement, either improvised or purposefully selected. This movement has aesthetic and often symbolic value. Dance can be categorized and described by its choreography, by its repertoi ...
database (2010)
* Epigenesis (biology)
* Epigenetics in forensic science
Epigenetics in forensic science is the application of epigenetics to solving crimes.
Forensic science has been using DNA as evidence since 1984, however this does not give information about any changes in the individual since birth and will not be ...
* Epigenetic therapy
Epigenetic therapy is the use of drugs or other epigenome-influencing techniques to treat medical conditions. Many diseases, including cancer, heart disease, diabetes, and mental illnesses are influenced by epigenetic mechanisms. Epigenetic th ...
* Epigenetics of neurodegenerative diseases
file:Parasagittal_MRI_of_human_head_in_patient_with_benign_familial_macrocephaly_prior_to_brain_injury_(ANIMATED).gif, Para-sagittal MRI of the head in a patient with benign familial macrocephaly.Neurodegenerative diseases are a heterogeneous grou ...
* Genetics
* Lamarckism
* Nutriepigenomics
* Position-effect variegation
* Preformationism
* Somatic epitype A somatic epitype is a non-heritable epigenetic alteration in a gene. It is similar to conventional epigenetics in that it does not involve changes in the DNA primary sequence. Physically, the somatic epitype corresponds to changes in DNA methyla ...
* Synthetic genetic array Synthetic genetic array analysis (SGA) is a high-throughput screening, high-throughput technique for exploring synthetic lethality, synthetic lethal and synthetic sick genetic interactions (Synthetic lethality, SSL). SGA allows for the systematic co ...
* Transcriptional memory
Transcriptional memory is a biological phenomenon, initially discovered in yeast, during which cells primed with a particular cue show increased rates of gene expression after re-stimulation at a later time. This event was shown to take place: in ...
* Transgenerational epigenetic inheritance
Transgenerational epigenetic inheritance is the transmission of epigenetic markers from one organism to the next (i.e., from parent to child) that affects the traits of offspring without altering the nucleic acid primary structure, primary structur ...
References
Further reading
*
External links
*
The Human Epigenome Project (HEP)
The Epigenome Network of Excellence (NoE)
Canadian Epigenetics, Environment and Health Research Consortium (CEEHRC)
The Epigenome Network of Excellence (NoE) – public international site
"DNA Is Not Destiny"
– ''Discover'' magazine cover story
"The Ghost In Your Genes"
''Horizon'' (2005), BBC
at Hopkins Medicine
{{Authority control
Genetic mapping
Lamarckism
 In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the
In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the  Because the phenotype of a cell or individual is affected by which of its genes are transcribed, heritable transcription states can give rise to epigenetic effects. There are several layers of regulation of
Because the phenotype of a cell or individual is affected by which of its genes are transcribed, heritable transcription states can give rise to epigenetic effects. There are several layers of regulation of  Although histone modifications occur throughout the entire sequence, the unstructured N-termini of histones (called histone tails) are particularly highly modified. These modifications include
Although histone modifications occur throughout the entire sequence, the unstructured N-termini of histones (called histone tails) are particularly highly modified. These modifications include  Areas of the brain important in the formation of memories include the hippocampus, medial prefrontal cortex (mPFC), anterior cingulate cortex and amygdala, as shown in the diagram of the human brain in this section.
When a strong memory is created, as in a rat subjected to contextual fear conditioning (CFC), one of the earliest events to occur is that more than 100 DNA double-strand breaks are formed by topoisomerase IIB in neurons of the hippocampus and the medial prefrontal cortex (mPFC). These double-strand breaks are at specific locations that allow activation of transcription of
Areas of the brain important in the formation of memories include the hippocampus, medial prefrontal cortex (mPFC), anterior cingulate cortex and amygdala, as shown in the diagram of the human brain in this section.
When a strong memory is created, as in a rat subjected to contextual fear conditioning (CFC), one of the earliest events to occur is that more than 100 DNA double-strand breaks are formed by topoisomerase IIB in neurons of the hippocampus and the medial prefrontal cortex (mPFC). These double-strand breaks are at specific locations that allow activation of transcription of  The DNA methyltransferases DNMT3A1, DNMT3A2 and DNMT3B can all methylate cytosines (see image this section) at CpG sites in or near the promoters of genes. As shown by Manzo et al., these three DNA methyltransferases differ in their genomic binding locations and DNA methylation activity at different regulatory sites. Manzo et al. located 3,970 genome regions exclusively enriched for DNMT3A1, 3,838 regions for DNMT3A2 and 3,432 regions for DNMT3B. When DNMT3A2 is newly induced as an IEG (when neurons are activated), many new cytosine methylations occur, presumably in the target regions of DNMT3A2. Oliviera et al. found that the neuronal activity-inducible IEG levels of Dnmt3a2 in the hippocampus determined the ability to form long-term memories.
Rats form long-term associative memories after contextual fear conditioning (CFC). Duke et al. found that 24 hours after CFC in rats, in hippocampus neurons, 2,097 genes (9.17% of the genes in the rat genome) had altered methylation. When newly methylated cytosines are present in CpG sites in the promoter regions of genes, the genes are often repressed, and when newly demethylated cytosines are present the genes may be activated. After CFC, there were 1,048 genes with reduced mRNA expression and 564 genes with upregulated mRNA expression. Similarly, when mice undergo CFC, one hour later in the hippocampus region of the mouse brain there are 675 demethylated genes and 613 hypermethylated genes. However, memories do not remain in the hippocampus, but after four or five weeks the memories are stored in the anterior cingulate cortex. In the studies on mice after CFC, Halder et al. showed that four weeks after CFC there were at least 1,000 differentially methylated genes and more than 1,000 differentially expressed genes in the anterior cingulate cortex, while at the same time the altered methylations in the hippocampus were reversed.
The epigenetic alteration of methylation after a new memory is established creates a different pool of nuclear mRNAs. As reviewed by Bernstein, the epigenetically determined new mix of nuclear
The DNA methyltransferases DNMT3A1, DNMT3A2 and DNMT3B can all methylate cytosines (see image this section) at CpG sites in or near the promoters of genes. As shown by Manzo et al., these three DNA methyltransferases differ in their genomic binding locations and DNA methylation activity at different regulatory sites. Manzo et al. located 3,970 genome regions exclusively enriched for DNMT3A1, 3,838 regions for DNMT3A2 and 3,432 regions for DNMT3B. When DNMT3A2 is newly induced as an IEG (when neurons are activated), many new cytosine methylations occur, presumably in the target regions of DNMT3A2. Oliviera et al. found that the neuronal activity-inducible IEG levels of Dnmt3a2 in the hippocampus determined the ability to form long-term memories.
Rats form long-term associative memories after contextual fear conditioning (CFC). Duke et al. found that 24 hours after CFC in rats, in hippocampus neurons, 2,097 genes (9.17% of the genes in the rat genome) had altered methylation. When newly methylated cytosines are present in CpG sites in the promoter regions of genes, the genes are often repressed, and when newly demethylated cytosines are present the genes may be activated. After CFC, there were 1,048 genes with reduced mRNA expression and 564 genes with upregulated mRNA expression. Similarly, when mice undergo CFC, one hour later in the hippocampus region of the mouse brain there are 675 demethylated genes and 613 hypermethylated genes. However, memories do not remain in the hippocampus, but after four or five weeks the memories are stored in the anterior cingulate cortex. In the studies on mice after CFC, Halder et al. showed that four weeks after CFC there were at least 1,000 differentially methylated genes and more than 1,000 differentially expressed genes in the anterior cingulate cortex, while at the same time the altered methylations in the hippocampus were reversed.
The epigenetic alteration of methylation after a new memory is established creates a different pool of nuclear mRNAs. As reviewed by Bernstein, the epigenetically determined new mix of nuclear  While epigenetics is of fundamental importance in
While epigenetics is of fundamental importance in