|
ChIP-seq
ChIP-sequencing, also known as ChIP-seq, is a method used to analyze protein interactions with DNA. ChIP-seq combines chromatin immunoprecipitation (ChIP) with Massively parallel signature sequencing, massively parallel DNA sequencing to identify the binding sites of DNA-associated proteins. It can be used to map global binding sites precisely for any protein of interest. Previously, ChIP-on-chip was the most common technique utilized to study these protein–DNA relations. Uses ChIP-seq is primarily used to determine how transcription factors and other chromatin-associated proteins influence phenotype-affecting mechanisms. Determining how proteins interact with DNA to regulate gene expression is essential for fully understanding many biological processes and disease states. This epigenetic information is complementary to genotype and expression analysis. ChIP-seq technology is currently seen primarily as an alternative to ChIP-chip which requires a protein microarray, hybridization ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) is a type of immunoprecipitation experimental technique used to investigate the interaction between proteins and DNA in the cell. It aims to determine whether specific proteins are associated with specific genomic regions, such as transcription factors on promoters or other DNA binding sites, and possibly define cistromes. ChIP also aims to determine the specific location in the genome that various histone modifications are associated with, indicating the target of the histone modifiers. ChIP is crucial for the advancements in the field of epigenomics and learning more about epigenetic phenomena. Briefly, the conventional method is as follows: # DNA and associated proteins on chromatin in living cells or tissues are crosslinked (this step is omitted in Native ChIP). # The DNA-protein complexes (chromatin-protein) are then sheared into ~500 bp DNA fragments by sonication or nuclease digestion. # Cross-linked DNA fragments associated with the prot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epigenetic
In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the DNA sequence. The Greek prefix '' epi-'' ( "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in addition to" the traditional genetic basis for inheritance. Epigenetics most often involves changes that affect the regulation of gene expression, but the term can also be used to describe any heritable phenotypic change. Such effects on cellular and physiological phenotypic traits may result from external or environmental factors, or be part of normal development. The term also refers to the mechanism of changes: functionally relevant alterations to the genome that do not involve mutation of the nucleotide sequence. Examples of mechanisms that produce such changes are DNA methylation and histone modification, each of which alters how genes are expressed without altering the underlying DNA sequence. Gene expression can ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ChIP
Chromatin immunoprecipitation (ChIP) is a type of immunoprecipitation experimental technique used to investigate the interaction between proteins and DNA in the cell. It aims to determine whether specific proteins are associated with specific genomic regions, such as transcription factors on promoter (biology), promoters or other DNA binding sites, and possibly define cistromes. ChIP also aims to determine the specific location in the genome that various histone modifications are associated with, indicating the target of the histone modifiers. ChIP is crucial for the advancements in the field of epigenomics and learning more about epigenetic phenomena. Briefly, the conventional method is as follows: # DNA and associated proteins on chromatin in living cells or tissues are crosslinked (this step is omitted in Native ChIP). # The DNA-protein complexes (chromatin-protein) are then sheared into ~500 bp DNA fragments by sonication or nuclease digestion. # Crosslinking of DNA, Cross-linked ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FAIRE-Seq
FAIRE-Seq (Formaldehyde-Assisted Isolation of Regulatory Elements) is a method in molecular biology used for determining the sequences of DNA regions in the genome associated with regulatory activity. The technique was developed in the laboratory of Jason D. Lieb at the University of North Carolina, Chapel Hill. In contrast to DNase-Seq, the FAIRE-Seq protocol doesn't require the permeabilization of cells or isolation of nuclei, and can analyse any cell type. In a study of seven diverse human cell types, DNase-seq and FAIRE-seq produced strong cross-validation, with each cell type having 1-2% of the human genome as open chromatin. Workflow The protocol is based on the fact that the formaldehyde cross-linking is more efficient in nucleosome-bound DNA than it is in nucleosome-depleted regions of the genome. This method then segregates the non cross-linked DNA that is usually found in open chromatin, which is then sequenced. The protocol consists of cross linking, phenol extraction a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residue ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cluster Amplification
may refer to: Science and technology Astronomy * Cluster (spacecraft), constellation of four European Space Agency spacecraft * Asteroid cluster, a small asteroid family * Cluster II (spacecraft), a European Space Agency mission to study the magnetosphere * Galaxy cluster, large gravitationally bound groups of galaxies, or groups of groups of galaxies * Supercluster, the largest gravitationally bound objects in the universe, composed of many galaxy clusters * Star cluster ** Globular cluster, a spherical collection of stars whose orbit is either partially or completely in the halo of the parent galaxy ** Open cluster, a spherical collection of stars that orbits a galaxy in the galactic plane Biology and medicine * Cancer cluster, in biomedicine, an occurrence of a greater-than-expected number of cancer cases * Cluster headache, a neurological disease that involves an immense degree of pain * Cluster of differentiation, protocol used for the identification and investigation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tiling Array
Tiling arrays are a subtype of microarray chips. Like traditional microarrays, they function by hybridizing labeled DNA or RNA target molecules to probes fixed onto a solid surface. Tiling arrays differ from traditional microarrays in the nature of the probes. Instead of probing for sequences of known or predicted genes that may be dispersed throughout the genome, tiling arrays probe intensively for sequences which are known to exist in a contiguous region. This is useful for characterizing regions that are sequenced, but whose local functions are largely unknown. Tiling arrays aid in transcriptome mapping as well as in discovering sites of DNA/protein interaction (ChIP-chip, DamID), of DNA methylation (MeDIP-chip) and of sensitivity to DNase (DNase Chip) and array CGH. In addition to detecting previously unidentified genes and regulatory sequences, improved quantification of transcription products is possible. Specific probes are present in millions of copies (as opposed to on ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small bits of nucleic acids can be manufactured as single-stranded molecules with any user-specified sequence, and so are vital for artificial gene synthesis, polymerase chain reaction (PCR), DNA sequencing, molecular cloning and as molecular probes. In nature, oligonucleotides are usually found as small RNA molecules that function in the regulation of gene expression (e.g. microRNA), or are degradation intermediates derived from the breakdown of larger nucleic acid molecules. Oligonucleotides are characterized by the sequence of nucleotide residues that make up the entire molecule. The length of the oligonucleotide is usually denoted by " -mer" (from Greek ''meros'', "part"). For example, an oligonucleotide of six nucleotides (nt) is a hexamer, while one of 25 nt wou ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
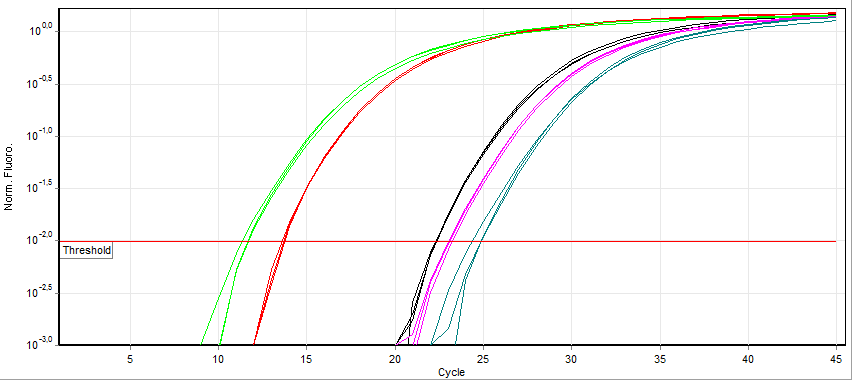
Real-time Polymerase Chain Reaction
A real-time polymerase chain reaction (real-time PCR, or qPCR) is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR (i.e., in real time), not at its end, as in conventional PCR. Real-time PCR can be used quantitatively (quantitative real-time PCR) and semi-quantitatively (i.e., above/below a certain amount of DNA molecules) (semi-quantitative real-time PCR). Two common methods for the detection of PCR products in real-time PCR are (1) non-specific fluorescent dyes that intercalate with any double-stranded DNA and (2) sequence-specific DNA probes consisting of oligonucleotides that are labelled with a fluorescent reporter, which permits detection only after hybridization of the probe with its complementary sequence. The Minimum Information for Publication of Quantitative Real-Time PCR Experiments ( MIQE) guidelines propose that the abbreviation ''qPCR'' be used for ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antibody
An antibody (Ab), also known as an immunoglobulin (Ig), is a large, Y-shaped protein used by the immune system to identify and neutralize foreign objects such as pathogenic bacteria and viruses. The antibody recognizes a unique molecule of the pathogen, called an antigen. Each tip of the "Y" of an antibody contains a paratope (analogous to a lock) that is specific for one particular epitope (analogous to a key) on an antigen, allowing these two structures to bind together with precision. Using this binding mechanism, an antibody can ''tag'' a microbe or an infected cell for attack by other parts of the immune system, or can neutralize it directly (for example, by blocking a part of a virus that is essential for its invasion). To allow the immune system to recognize millions of different antigens, the antigen-binding sites at both tips of the antibody come in an equally wide variety. In contrast, the remainder of the antibody is relatively constant. It only occurs in a few varia ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Crosslinking Of DNA
In genetics, crosslinking of DNA occurs when various exogenous or endogenous agents react with two nucleotides of DNA, forming a covalent linkage between them. This crosslink can occur within the same strand (intrastrand) or between opposite strands of double-stranded DNA (interstrand). These adducts interfere with cellular metabolism, such as DNA replication and transcription, triggering cell death. These crosslinks can, however, be repaired through excision or recombination pathways. DNA crosslinking also has useful merit in chemotherapy and targeting cancerous cells for apoptosis, as well as in understanding how proteins interact with DNA. Crosslinking agents Many characterized crosslinking agents have two independently reactive groups within the same molecule, each of which is able to bind with a nucleotide residue of DNA. These agents are separated based upon their source of origin and labeled either as exogenous or endogenous. Exogenous crosslinking agents are chemicals an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cell (biology)
The cell is the basic structural and functional unit of life forms. Every cell consists of a cytoplasm enclosed within a membrane, and contains many biomolecules such as proteins, DNA and RNA, as well as many small molecules of nutrients and metabolites.Cell Movements and the Shaping of the Vertebrate Body in Chapter 21 of Molecular Biology of the Cell '' fourth edition, edited by Bruce Alberts (2002) published by Garland Science. The Alberts text discusses how the "cellular building blocks" move to shape developing embryos. It is also common to describe small molecules such as ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |