|
Glycylpeptide N-tetradecanoyltransferase
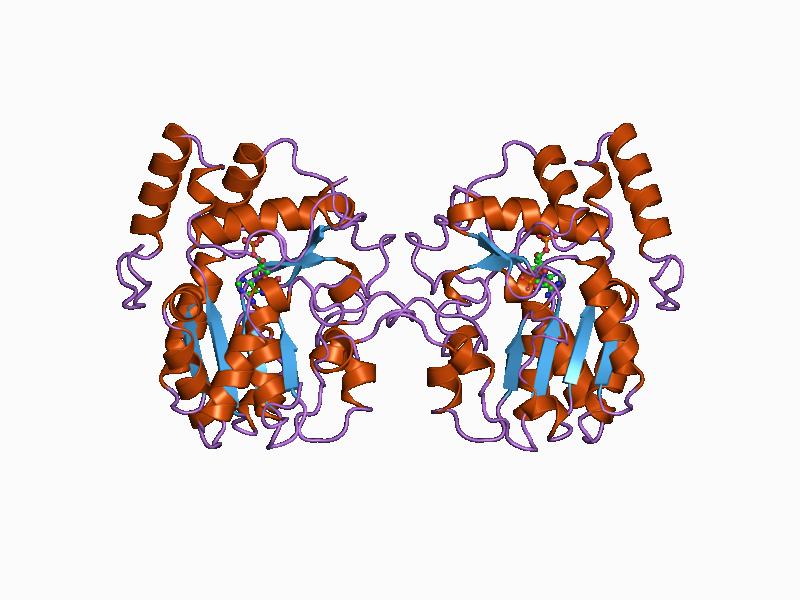
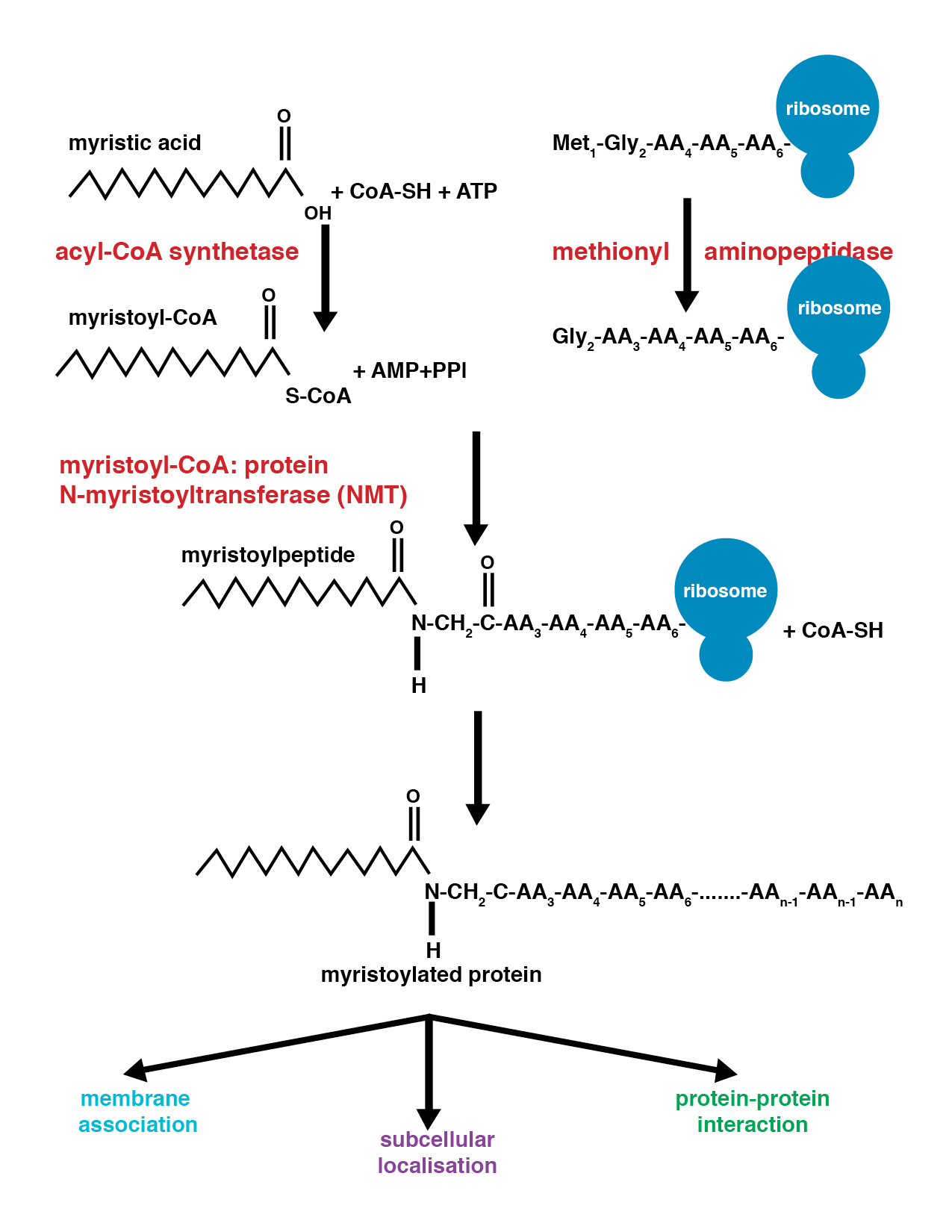
In enzymology, a glycylpeptide N-tetradecanoyltransferase () is an enzyme that catalyzes the chemical reaction :tetradecanoyl-CoA + glycylpeptide \rightleftharpoons CoA + N-tetradecanoylglycylpeptide Thus, the two substrates of this enzyme are tetradecanoyl-CoA and glycylpeptide, whereas its two products are CoA and N-tetradecanoylglycylpeptide. It participates in the N-Myristoylation of proteins, and in vertebrates there are two isoenzymes NMT1 and NMT2. Besides tetradecanoyl-CoA, this enzyme is also capable of using modified versions of this substrate. In human retina, an even wider range of fatty acids, including 14:1 n–9, 14:2n–6, and 12:0, are accepted by the enzyme and grafted onto guanylate cyclase activators. This is mainly a result of a special set of fatty-acid-CoA substrates available in the retina. Nomenclature This enzyme belongs to the family of transferases, specifically those N-acyltransferases transferring groups other than aminoacyl groups (cd0430 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzymology
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reacti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
N-myristoyltransferase 1
Glycylpeptide N-tetradecanoyltransferase 1 also known as myristoyl-CoA:protein N-myristoyltransferase 1 (NMT-1) is an enzyme that in humans is encoded by the ''NMT1'' gene. It belongs to the protein N-terminal methyltransferase and glycylpeptide N-tetradecanoyltransferase family of enzymes. References Further reading * * * * * * * * * * * * * * * * * * * See also * Myristoylation Myristoylation is a lipidation modification where a myristoyl group, derived from myristic acid, is covalently attached by an amide bond to the alpha-amino group of an N-terminus, N-terminal glycine residue. Myristic acid is a 14-carbon saturat ... * N-myristoyltransferase 2 External links * EC 2.3.1 Human proteins {{gene-17-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EF-hand
The EF hand is a helix–loop–helix structural domain or ''motif'' found in a large family of calcium-binding proteins. The EF-hand motif contains a helix–loop–helix topology, much like the spread thumb and forefinger of the human hand, in which the Ca2+ ions are coordinated by ligands within the loop. The motif takes its name from traditional nomenclature used in describing the protein parvalbumin, which contains three such motifs and is probably involved in muscle relaxation via its calcium-binding activity. The EF-hand consists of two alpha helices linked by a short loop region (usually about 12 amino acids) that usually binds calcium ions. EF-hands also appear in each structural domain of the signaling protein calmodulin and in the muscle protein troponin-C. Calcium ion binding site The calcium ion is coordinated in a pentagonal bipyramidal configuration. The six residues involved in the binding are in positions 1, 3, 5, 7, 9 and 12; these residues are denoted by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Data Bank
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules, such as proteins and nucleic acids. The data, typically obtained by X-ray crystallography, NMR spectroscopy, or, increasingly, cryo-electron microscopy, and submitted by biologists and biochemists from around the world, are freely accessible on the Internet via the websites of its member organisations (PDBe, PDBj, RCSB, and BMRB). The PDB is overseen by an organization called the Worldwide Protein Data Bank, wwPDB. The PDB is a key in areas of structural biology, such as structural genomics. Most major scientific journals and some funding agencies now require scientists to submit their structure data to the PDB. Many other databases use protein structures deposited in the PDB. For example, SCOP and CATH classify protein structures, while PDBsum provides a graphic overview of PDB entries using information from other sources, such as Gene ontology. History Two ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tertiary Structure
Protein tertiary structure is the three dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains may interact and bond in a number of ways. The interactions and bonds of side chains within a particular protein determine its tertiary structure. The protein tertiary structure is defined by its atomic coordinates. These coordinates may refer either to a protein domain or to the entire tertiary structure.Branden C. and Tooze J. "Introduction to Protein Structure" Garland Publishing, New York. 1990 and 1991. A number of tertiary structures may fold into a quaternary structure.Kyte, J. "Structure in Protein Chemistry." Garland Publishing, New York. 1995. History The science of the tertiary structure of proteins has progressed from one of hypothesis to one of detailed definition. Although Emil Fischer had suggested proteins were made of poly ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Enzymes
This article lists enzymes by their classification in the International Union of Biochemistry and Molecular Biology's Enzyme Commission (EC) numbering system. * List of EC numbers (EC 5) * List of EC numbers (EC 6) :Oxidoreductases (EC 1) ( Oxidoreductase) * Dehydrogenase *Luciferase * DMSO reductase :EC 1.1 (act on the CH-OH group of donors) * :EC 1.1.1 (with NAD+ or NADP+ as acceptor) ** Alcohol dehydrogenase (NAD) ** Alcohol dehydrogenase (NADP) ** Homoserine dehydrogenase ** Aminopropanol oxidoreductase **Diacetyl reductase ** Glycerol dehydrogenase ** Propanediol-phosphate dehydrogenase **glycerol-3-phosphate dehydrogenase (NAD+) **D-xylulose reductase ** L-xylulose reductase ** Lactate dehydrogenase ** Malate dehydrogenase ** Isocitrate dehydrogenase **HMG-CoA reductase * :EC 1.1.2 (with a cytochrome as acceptor) * :EC 1.1.3 (with oxygen as acceptor) ** Glucose oxidase ** L-gulonolactone oxidase ** Thiamine oxidase ** Xanthine oxidase * :EC 1.1. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Acyltransferase
Acyltransferase is a type of transferase enzyme that acts upon acyl groups. Examples include: * Glyceronephosphate O-acyltransferase * Lecithin-cholesterol acyltransferase *Long-chain-alcohol O-fatty-acyltransferase In enzymology, a long-chain-alcohol O-fatty-acyltransferase () is an enzyme that catalyzes the chemical reaction :acyl-CoA + a long-chain alcohol \rightleftharpoons CoA + a long-chain ester Thus, the two substrates of this enzyme are acyl-Co ... See also * Acetyltransferase External links * Transferases EC 2.3 {{2.3-enzyme-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transferase
A transferase is any one of a class of enzymes that catalyse the transfer of specific functional groups (e.g. a methyl or glycosyl group) from one molecule (called the donor) to another (called the acceptor). They are involved in hundreds of different biochemical pathways throughout biology, and are integral to some of life's most important processes. Transferases are involved in myriad reactions in the cell. Three examples of these reactions are the activity of coenzyme A (CoA) transferase, which transfers thiol esters, the action of N-acetyltransferase, which is part of the pathway that metabolizes tryptophan, and the regulation of pyruvate dehydrogenase (PDH), which converts pyruvate to acetyl CoA. Transferases are also utilized during translation. In this case, an amino acid chain is the functional group transferred by a peptidyl transferase. The transfer involves the removal of the growing amino acid chain from the tRNA molecule in the A-site of the ribosome a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Guanylate Cyclase Activator
A guanylate cyclase activator (or "GUCA") is one of group of proteins which upregulates guanylate cyclase. It is also known as guanylate cyclase-activating protein, with the abbreviation "GCAP". Mutations In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mitosi ... can be associated with vision defects. There are five genes involved: * , , * , References External links * Proteins {{biochemistry-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NMT2
Glycylpeptide N-tetradecanoyltransferase 2 known also as N-myristoyltransferase, is an enzyme (EC: 2.3.1.97) that in humans is encoded by the ''NMT2'' gene. Function N-myristoyltransferase (NMT) catalyzes the reaction of N-terminal myristoylation of many signaling proteins. It transfers myristic acid from myristoyl coenzyme A to the amino group of a protein's N-terminal glycine residue. Biochemical evidence indicates the presence of several distinct NMTs, varying in apparent molecular weight and /or subcellular distribution. The 496-amino acid of human NMT2 protein shares 77% and 96% sequence identity with human NMT1 and mouse Nmt2 comprise two distinct families of N-myristoyltransferases. Interactions NMT2 has been shown to interact with: * caspase 3 * MARCKS See also * N-myristoyltransferase 1 Glycylpeptide N-tetradecanoyltransferase 1 also known as myristoyl-CoA:protein N-myristoyltransferase 1 (NMT-1) is an enzyme that in humans is encoded by the ''NMT1'' gene. It b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Myristoylation
Myristoylation is a lipidation modification where a myristoyl group, derived from myristic acid, is covalently attached by an amide bond to the alpha-amino group of an N-terminal glycine residue. Myristic acid is a 14-carbon saturated fatty acid (14:0) with the systematic name of ''n''-Tetradecanoic acid. This modification can be added either co-translationally or post-translationally. N-myristoyltransferase (NMT) catalyzes the myristic acid addition reaction in the cytoplasm of cells. This lipidation event is the most found type of fatty acylation and is common among many organisms including animals, plants, fungi, protozoans and viruses. Myristoylation allows for weak protein–protein and protein–lipid interactions and plays an essential role in membrane targeting, protein–protein interactions and functions widely in a variety of signal transduction pathways. Discovery In 1982, Koiti Titani's lab identified an "N-terminal blocking group" on the catalytic subunit of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different molecules known as product (chemistry), products. Almost all metabolism, metabolic processes in the cell (biology), cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme, pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are Ribozyme, catalytic RNA molecules, called ribozymes. Enzymes' Chemical specificity, specific ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |