Recombination Repair on:
[Wikipedia]
[Google]
[Amazon]
 Homologous recombination is a type of
Homologous recombination is a type of
 In the early 1900s,
In the early 1900s,
 Double-strand breaks can be repaired through homologous recombination, polymerase theta-mediated end joining (TMEJ) or through non-homologous end joining (NHEJ). NHEJ is a DNA repair mechanism which, unlike homologous recombination, does not require a long
Double-strand breaks can be repaired through homologous recombination, polymerase theta-mediated end joining (TMEJ) or through non-homologous end joining (NHEJ). NHEJ is a DNA repair mechanism which, unlike homologous recombination, does not require a long
 Two primary models for how homologous recombination repairs double-strand breaks in DNA are the double-strand break repair (DSBR) pathway (sometimes called the ''double Holliday junction model'') and the synthesis-dependent strand annealing (SDSA) pathway. The two pathways are similar in their first several steps. After a double-strand break occurs, the MRX complex (
Two primary models for how homologous recombination repairs double-strand breaks in DNA are the double-strand break repair (DSBR) pathway (sometimes called the ''double Holliday junction model'') and the synthesis-dependent strand annealing (SDSA) pathway. The two pathways are similar in their first several steps. After a double-strand break occurs, the MRX complex (
 After the stages of resection, strand invasion and DNA synthesis, the DSBR and SDSA pathways become distinct. The DSBR pathway is unique in that the second 3' overhang (which was not involved in strand invasion) also forms a Holliday junction with the homologous chromosome. The double Holliday junctions are then converted into recombination products by
After the stages of resection, strand invasion and DNA synthesis, the DSBR and SDSA pathways become distinct. The DSBR pathway is unique in that the second 3' overhang (which was not involved in strand invasion) also forms a Holliday junction with the homologous chromosome. The double Holliday junctions are then converted into recombination products by
 The single-strand annealing (SSA) pathway of homologous recombination repairs double-strand breaks between two repeat sequences. The SSA pathway is unique in that it does not require a separate similar or identical molecule of DNA, like the DSBR or SDSA pathways of homologous recombination. Instead, the SSA pathway only requires a single DNA duplex, and uses the repeat sequences as the identical sequences that homologous recombination needs for repair. The pathway is relatively simple in concept: after two strands of the same DNA duplex are cut back around the site of the double-strand break, the two resulting 3' overhangs then align and anneal to each other, restoring the DNA as a continuous duplex.
As DNA around the double-strand break is cut back, the single-stranded 3' overhangs being produced are coated with the RPA protein, which prevents the 3' overhangs from sticking to themselves. A protein called Rad52 then binds each of the repeat sequences on either side of the break, and aligns them to enable the two complementary repeat sequences to anneal. After annealing is complete, leftover non-homologous flaps of the 3' overhangs are cut away by a set of nucleases, known as Rad1/Rad10, which are brought to the flaps by the Saw1 and Slx4 proteins. New DNA synthesis fills in any gaps, and ligation restores the DNA duplex as two continuous strands. The DNA sequence between the repeats is always lost, as is one of the two repeats. The SSA pathway is considered
The single-strand annealing (SSA) pathway of homologous recombination repairs double-strand breaks between two repeat sequences. The SSA pathway is unique in that it does not require a separate similar or identical molecule of DNA, like the DSBR or SDSA pathways of homologous recombination. Instead, the SSA pathway only requires a single DNA duplex, and uses the repeat sequences as the identical sequences that homologous recombination needs for repair. The pathway is relatively simple in concept: after two strands of the same DNA duplex are cut back around the site of the double-strand break, the two resulting 3' overhangs then align and anneal to each other, restoring the DNA as a continuous duplex.
As DNA around the double-strand break is cut back, the single-stranded 3' overhangs being produced are coated with the RPA protein, which prevents the 3' overhangs from sticking to themselves. A protein called Rad52 then binds each of the repeat sequences on either side of the break, and aligns them to enable the two complementary repeat sequences to anneal. After annealing is complete, leftover non-homologous flaps of the 3' overhangs are cut away by a set of nucleases, known as Rad1/Rad10, which are brought to the flaps by the Saw1 and Slx4 proteins. New DNA synthesis fills in any gaps, and ligation restores the DNA duplex as two continuous strands. The DNA sequence between the repeats is always lost, as is one of the two repeats. The SSA pathway is considered
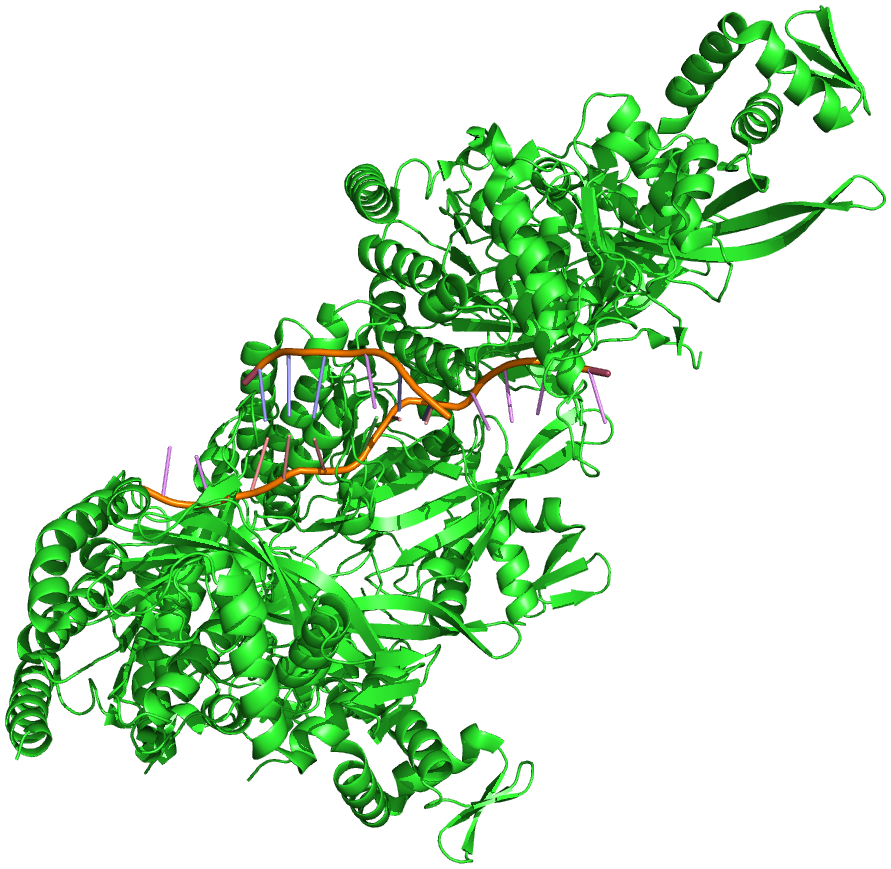 Homologous recombination is a major DNA repair process in bacteria. It is also important for producing genetic diversity in bacterial populations, although the process differs substantially from
Homologous recombination is a major DNA repair process in bacteria. It is also important for producing genetic diversity in bacterial populations, although the process differs substantially from
 The SARS-CoV-2 outbreak in Diamond Princess cruise most likely originated from either a single person infected with a virus variant identical to the Wuhan WIV04 isolates, or simultaneously with another primary case infected with a virus containing the 11083G > T mutation. Linkage disequilibrium analysis confirmed that
The SARS-CoV-2 outbreak in Diamond Princess cruise most likely originated from either a single person infected with a virus variant identical to the Wuhan WIV04 isolates, or simultaneously with another primary case infected with a virus containing the 11083G > T mutation. Linkage disequilibrium analysis confirmed that
 Without proper homologous recombination, chromosomes often incorrectly align for the first phase of cell division in meiosis. This causes chromosomes to fail to properly segregate in a process called nondisjunction. In turn, nondisjunction can cause
Without proper homologous recombination, chromosomes often incorrectly align for the first phase of cell division in meiosis. This causes chromosomes to fail to properly segregate in a process called nondisjunction. In turn, nondisjunction can cause
 While the pathways can mechanistically vary, the ability of organisms to perform homologous recombination is universally conserved across all domains of life. Based on the similarity of their amino acid sequences, homologs of a number of proteins can be found in multiple domains of life indicating that they evolved a long time ago, and have since diverged from common ancestral proteins.
RecA recombinase family members are found in almost all organisms with RecA in bacteria,
While the pathways can mechanistically vary, the ability of organisms to perform homologous recombination is universally conserved across all domains of life. Based on the similarity of their amino acid sequences, homologs of a number of proteins can be found in multiple domains of life indicating that they evolved a long time ago, and have since diverged from common ancestral proteins.
RecA recombinase family members are found in almost all organisms with RecA in bacteria,
 Many methods for introducing DNA sequences into organisms to create recombinant DNA and
Many methods for introducing DNA sequences into organisms to create recombinant DNA and
Animations – homologous recombination
Animations showing several models of homologous recombination
Animation of the bacterial RecBCD pathway of homologous recombination {{DEFAULTSORT:Homologous Recombination DNA repair Modification of genetic information Telomeres
genetic recombination
Genetic recombination (also known as genetic reshuffling) is the exchange of genetic material between different organisms which leads to production of offspring with combinations of traits that differ from those found in either parent. In eukaryo ...
in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acid
Nucleic acids are biopolymers, macromolecules, essential to all known forms of life. They are composed of nucleotides, which are the monomers made of three components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main cl ...
s (usually DNA as in cellular organisms but may be also RNA in viruses).
Homologous recombination is widely used by cells to accurately DNA repair harmful breaks that occur on both strands of DNA, known as double-strand breaks (DSB), in a process called homologous recombinational repair (HRR).
Homologous recombination also produces new combinations of DNA sequences during meiosis, the process by which eukaryote
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacte ...
s make gamete cells, like sperm
Sperm is the male reproductive cell, or gamete, in anisogamous forms of sexual reproduction (forms in which there is a larger, female reproductive cell and a smaller, male one). Animals produce motile sperm with a tail known as a flagellum, whi ...
and egg cells in animals. These new combinations of DNA represent genetic variation
Genetic variation is the difference in DNA among individuals or the differences between populations. The multiple sources of genetic variation include mutation and genetic recombination. Mutations are the ultimate sources of genetic variation, ...
in offspring, which in turn enables populations to adapt during the course of evolution.
Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species of bacteria and viruses. Horizontal gene transfer is the primary mechanism for the spread of antibiotic resistance in bacteria.
Although homologous recombination varies widely among different organisms and cell types, for double-stranded DNA ( dsDNA) most forms involve the same basic steps. After a double-strand break occurs, sections of DNA around the 5' ends of the break are cut away in a process called '' resection''. In the ''strand invasion'' step that follows, an overhanging 3' end
Directionality, in molecular biology and biochemistry, is the end-to-end chemical orientation of a single strand of nucleic acid. In a single strand of DNA or RNA, the chemical convention of naming carbon atoms in the nucleotide pentose-sugar-ri ...
of the broken DNA molecule then "invades" a similar or identical DNA molecule that is not broken. After strand invasion, the further sequence of events may follow either of two main pathways discussed below (see Models); the DSBR (double-strand break repair) pathway or the SDSA (synthesis-dependent strand annealing) pathway. Homologous recombination that occurs during DNA repair tends to result in non-crossover products, in effect restoring the damaged DNA molecule as it existed before the double-strand break.
Homologous recombination is conserved across all three domains of life as well as DNA and RNA viruses, suggesting that it is a nearly universal biological mechanism. The discovery of genes for homologous recombination in protists—a diverse group of eukaryotic microorganisms
A microorganism, or microbe,, ''mikros'', "small") and ''organism'' from the el, ὀργανισμός, ''organismós'', "organism"). It is usually written as a single word but is sometimes hyphenated (''micro-organism''), especially in olde ...
—has been interpreted as evidence that meiosis emerged early in the evolution of eukaryotes. Since their dysfunction has been strongly associated with increased susceptibility to several types of cancer, the proteins that facilitate homologous recombination are topics of active research. Homologous recombination is also used in gene targeting, a technique for introducing genetic changes into target organisms. For their development of this technique, Mario Capecchi, Martin Evans and Oliver Smithies were awarded the 2007 Nobel Prize for Physiology or Medicine; Capecchi and Smithies independently discovered applications to mouse embryonic stem cells, however the highly conserved mechanisms underlying the DSB repair model, including uniform homologous integration of transformed DNA (gene therapy), were first shown in plasmid experiments by Orr-Weaver, Szostack and Rothstein. Researching the plasmid-induced DSB, using γ-irradiation in the 1970s-1980s, led to later experiments using endonucleases (e.g. I-SceI) to cut chromosomes for genetic engineering of mammalian cells, where nonhomologous recombination is more frequent than in yeast.
History and discovery
 In the early 1900s,
In the early 1900s, William Bateson
William Bateson (8 August 1861 – 8 February 1926) was an English biologist who was the first person to use the term genetics to describe the study of heredity, and the chief populariser of the ideas of Gregor Mendel following their rediscover ...
and Reginald Punnett found an exception to one of the principles of inheritance originally described by Gregor Mendel in the 1860s. In contrast to Mendel's notion that traits are independently assorted when passed from parent to child—for example that a cat's hair color and its tail length are inherited independent of each other—Bateson and Punnett showed that certain genes associated with physical traits can be inherited together, or genetically linked. In 1911, after observing that linked traits could on occasion be inherited separately, Thomas Hunt Morgan suggested that " crossovers" can occur between linked genes, where one of the linked genes physically crosses over to a different chromosome. Two decades later, Barbara McClintock
Barbara McClintock (June 16, 1902 – September 2, 1992) was an American scientist and cytogeneticist who was awarded the 1983 Nobel Prize in Physiology or Medicine. McClintock received her PhD in botany from Cornell University in 1927. There s ...
and Harriet Creighton demonstrated that chromosomal crossover occurs during meiosis, the process of cell division by which sperm
Sperm is the male reproductive cell, or gamete, in anisogamous forms of sexual reproduction (forms in which there is a larger, female reproductive cell and a smaller, male one). Animals produce motile sperm with a tail known as a flagellum, whi ...
and egg cells are made. Within the same year as McClintock's discovery, Curt Stern showed that crossing over—later called "recombination"—could also occur in somatic cell
A somatic cell (from Ancient Greek σῶμα ''sôma'', meaning "body"), or vegetal cell, is any biological cell forming the body of a multicellular organism other than a gamete, germ cell, gametocyte or undifferentiated stem cell. Such cells compo ...
s like white blood cell
White blood cells, also called leukocytes or leucocytes, are the cell (biology), cells of the immune system that are involved in protecting the body against both infectious disease and foreign invaders. All white blood cells are produced and de ...
s and skin cells that divide through mitosis
In cell biology, mitosis () is a part of the cell cycle in which replicated chromosomes are separated into two new nuclei. Cell division by mitosis gives rise to genetically identical cells in which the total number of chromosomes is mainta ...
.
In 1947, the microbiologist Joshua Lederberg showed that bacteria—which had been assumed to reproduce only asexually through binary fission—are capable of genetic recombination, which is more similar to sexual reproduction. This work established ''E. coli
''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus ''Escher ...
'' as a model organism
A model organism (often shortened to model) is a non-human species that is extensively studied to understand particular biological phenomena, with the expectation that discoveries made in the model organism will provide insight into the workin ...
in genetics, and helped Lederberg win the 1958 Nobel Prize in Physiology or Medicine. Building on studies in fungi, in 1964 Robin Holliday proposed a model for recombination in meiosis which introduced key details of how the process can work, including the exchange of material between chromosomes through Holliday junction
A Holliday junction is a branched nucleic acid structure that contains four double-stranded arms joined. These arms may adopt one of several conformations depending on buffer salt concentrations and the sequence of nucleobases closest to the ju ...
s. In 1983, Jack Szostak and colleagues presented a model now known as the DSBR pathway, which accounted for observations not explained by the Holliday model. During the next decade, experiments in '' Drosophila'', budding yeast and mammalian cells led to the emergence of other models of homologous recombination, called SDSA pathways, which do not always rely on Holliday junctions.
Much of the later work identifying proteins involved in the process and determining their mechanisms has been performed by a number of individuals including James Haber
James Haber is an American molecular biologist who is known for his discoveries in the field of DNA repair, in particular for his contributions to understanding the mechanisms of non-homologous end joining and Microhomology-mediated end joining, a ...
, Patrick Sung
Patrick Sung is an American professor of structural biology and biochemistry at the University of Texas Health Science Center at San Antonio. He is known for his work on DNA repair.
Biography
Sung was born on May 24, 1959 in Hong Kong. In 1981, h ...
, Stephen Kowalczykowski
Stephen Charles Kowalczykowski ("Steve K") is a Distinguished Professor oMicrobiology and Molecular Geneticsat thUniversity of California at Davis His research focuses on the biochemistry and molecular biology of DNA repair and homologous recombina ...
, and others.
In eukaryotes
Homologous recombination (HR) is essential to cell division in eukaryotes like plants, animals, fungi and protists. In cells that divide throughmitosis
In cell biology, mitosis () is a part of the cell cycle in which replicated chromosomes are separated into two new nuclei. Cell division by mitosis gives rise to genetically identical cells in which the total number of chromosomes is mainta ...
, homologous recombination repairs double-strand breaks in DNA caused by ionizing radiation
Ionizing radiation (or ionising radiation), including nuclear radiation, consists of subatomic particles or electromagnetic waves that have sufficient energy to ionize atoms or molecules by detaching electrons from them. Some particles can travel ...
or DNA-damaging chemicals. Left unrepaired, these double-strand breaks can cause large-scale rearrangement of chromosomes in somatic cell
A somatic cell (from Ancient Greek σῶμα ''sôma'', meaning "body"), or vegetal cell, is any biological cell forming the body of a multicellular organism other than a gamete, germ cell, gametocyte or undifferentiated stem cell. Such cells compo ...
s, which can in turn lead to cancer.
In addition to repairing DNA, homologous recombination also helps produce genetic diversity
Genetic diversity is the total number of genetic characteristics in the genetic makeup of a species, it ranges widely from the number of species to differences within species and can be attributed to the span of survival for a species. It is dis ...
when cells divide in meiosis to become specialized gamete cells—sperm
Sperm is the male reproductive cell, or gamete, in anisogamous forms of sexual reproduction (forms in which there is a larger, female reproductive cell and a smaller, male one). Animals produce motile sperm with a tail known as a flagellum, whi ...
or egg cells in animals, pollen
Pollen is a powdery substance produced by seed plants. It consists of pollen grains (highly reduced microgametophytes), which produce male gametes (sperm cells). Pollen grains have a hard coat made of sporopollenin that protects the gametophyt ...
or ovules in plants, and spore
In biology, a spore is a unit of sexual or asexual reproduction that may be adapted for dispersal and for survival, often for extended periods of time, in unfavourable conditions. Spores form part of the life cycles of many plants, algae, f ...
s in fungi. It does so by facilitating chromosomal crossover
Chromosomal crossover, or crossing over, is the exchange of genetic material during sexual reproduction between two homologous chromosomes' non-sister chromatids that results in recombinant chromosomes. It is one of the final phases of geneti ...
, in which regions of similar but not identical DNA are exchanged between homologous chromosomes. This creates new, possibly beneficial combinations of genes, which can give offspring an evolutionary advantage. Chromosomal crossover often begins when a protein called Spo11
Spo11 is a protein that in humans is encoded by the ''SPO11'' gene. Spo11, in a complex with mTopVIB, creates double strand breaks to initiate meiotic recombination. Its active site contains a tyrosine which ligates and dissociates with DNA to pr ...
makes a targeted double-strand break in DNA. These sites are non-randomly located on the chromosomes; usually in intergenic promoter regions and preferentially in GC-rich domains These double-strand break sites often occur at recombination hotspots, regions in chromosomes that are about 1,000–2,000 base pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
s in length and have high rates of recombination. The absence of a recombination hotspot between two genes on the same chromosome often means that those genes will be inherited by future generations in equal proportion. This represents linkage
Linkage may refer to:
* ''Linkage'' (album), by J-pop singer Mami Kawada, released in 2010
*Linkage (graph theory), the maximum min-degree of any of its subgraphs
*Linkage (horse), an American Thoroughbred racehorse
* Linkage (hierarchical cluster ...
between the two genes greater than would be expected from genes that independently assort during meiosis.
Timing within the mitotic cell cycle
homologous
Homology may refer to:
Sciences
Biology
*Homology (biology), any characteristic of biological organisms that is derived from a common ancestor
*Sequence homology, biological homology between DNA, RNA, or protein sequences
* Homologous chrom ...
sequence to guide repair. Whether homologous recombination or NHEJ is used to repair double-strand breaks is largely determined by the phase of cell cycle. Homologous recombination repairs DNA before the cell enters mitosis (M phase). It occurs during and shortly after DNA replication, in the S and G2 phases of the cell cycle, when sister chromatids are more easily available. Compared to homologous chromosomes, which are similar to another chromosome but often have different alleles, sister chromatids are an ideal template for homologous recombination because they are an identical copy of a given chromosome. When no homologous template is available or when the template cannot be accessed due to a defect in homologous recombination, the break is repaired via TMEJ in the S and G2 phases of the cell cycle. In contrast to homologous recombination and TMEJ, NHEJ is predominant in the G1 phase of the cell cycle, when the cell is growing but not yet ready to divide. It occurs less frequently after the G1 phase, but maintains at least some activity throughout the cell cycle. The mechanisms that regulate homologous recombination and NHEJ throughout the cell cycle vary widely between species.
Cyclin-dependent kinase
Cyclin-dependent kinases (CDKs) are the families of protein kinases first discovered for their role in regulating the cell cycle. They are also involved in regulating transcription, mRNA processing, and the differentiation of nerve cells. They a ...
s (CDKs), which modify the activity of other proteins by adding phosphate groups to (that is, phosphorylating
In chemistry, phosphorylation is the attachment of a phosphate group to a molecule or an ion. This process and its inverse, dephosphorylation, are common in biology and could be driven by natural selection. Text was copied from this source, whi ...
) them, are important regulators of homologous recombination in eukaryotes. When DNA replication begins in budding yeast, the cyclin-dependent kinase Cdc28 begins homologous recombination by phosphorylating the Sae2 protein. After being so activated by the addition of a phosphate, Sae2 causes a clean cut to be made near a double-strand break in DNA. It is unclear if the endonuclease
Endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (without regard to sequence), while many, typically called restriction endonucleases ...
responsible for this cut is Sae2 itself or another protein, Mre11. This allows a protein complex including Mre11, known as the MRX complex The MRX complex is a heterotrimeric protein complex consisting of Mre11, Rad50, and Xrs2. It is a budding yeast homolog of the mammalian Mre11-Rad50-Nbs1 (MRN) DNA damage repair complex.
Double-strand break repair
Cells are able to accurately rep ...
, to bind to DNA, and begins a series of protein-driven reactions that exchange material between two DNA molecules.
The role of chromatin
The packaging of eukaryotic DNA into chromatin presents a barrier to all DNA-based processes that require recruitment of enzymes to their sites of action. To allow homologous recombination (HR) DNA repair, the chromatin must be remodeled. In eukaryotes, ATP dependentchromatin remodeling
Chromatin remodeling is the dynamic modification of chromatin architecture to allow access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby control gene expression. Such remodeling is principally carried out ...
complexes and histone-modifying enzymes are two predominant factors employed to accomplish this remodeling process.
Chromatin relaxation occurs rapidly at the site of a DNA damage. In one of the earliest steps, the stress-activated protein kinase, c-Jun N-terminal kinase (JNK), phosphorylates SIRT6 on serine 10 in response to double-strand breaks or other DNA damage. This post-translational modification facilitates the mobilization of SIRT6 to DNA damage sites, and is required for efficient recruitment of poly (ADP-ribose) polymerase 1 (PARP1) to DNA break sites and for efficient repair of DSBs. PARP1 protein starts to appear at DNA damage sites in less than a second, with half maximum accumulation within 1.6 seconds after the damage occurs. Next the chromatin remodeler Alc1
Chromodomain-helicase-DNA-binding protein 1-like (ALC1) is an enzyme that in humans is encoded by the ''CHD1L'' gene. It has been implicated in chromatin remodeling and DNA relaxation process required for DNA replication, repair and transcription. ...
quickly attaches to the product of PARP1 action, a poly-ADP ribose chain, and Alc1 completes arrival at the DNA damage within 10 seconds of the occurrence of the damage. About half of the maximum chromatin relaxation, presumably due to action of Alc1, occurs by 10 seconds. This then allows recruitment of the DNA repair enzyme MRE11, to initiate DNA repair, within 13 seconds.
γH2AX, the phosphorylated form of H2AX is also involved in the early steps leading to chromatin decondensation after DNA double-strand breaks. The histone variant H2AX constitutes about 10% of the H2A histones in human chromatin. γH2AX (H2AX phosphorylated on serine 139) can be detected as soon as 20 seconds after irradiation of cells (with DNA double-strand break formation), and half maximum accumulation of γH2AX occurs in one minute. The extent of chromatin with phosphorylated γH2AX is about two million base pairs at the site of a DNA double-strand break. γH2AX does not, itself, cause chromatin decondensation, but within 30 seconds of irradiation, RNF8 protein can be detected in association with γH2AX. RNF8 mediates extensive chromatin decondensation, through its subsequent interaction with CHD4, a component of the nucleosome remodeling and deacetylase complex NuRD.
After undergoing relaxation subsequent to DNA damage, followed by DNA repair, chromatin recovers to a compaction state close to its pre-damage level after about 20 min.
Homologous recombination during meiosis
In vertebrates the locations at which recombination occurs are determined by the binding locations of PRDM9, a protein which recognizes a specific sequence motif by its zinc finger array. At these sites, another protein,SPO11
Spo11 is a protein that in humans is encoded by the ''SPO11'' gene. Spo11, in a complex with mTopVIB, creates double strand breaks to initiate meiotic recombination. Its active site contains a tyrosine which ligates and dissociates with DNA to pr ...
catalyses recombination-initiating double strand breaks (DSBs), a subset of which are repaired by recombination with the homologous chromosome. PRDM9 deposits both H3K4me3 and H3K36me3 histone methylation
Histone methylation is a process by which methyl groups are transferred to amino acids of histone proteins that make up nucleosomes, which the DNA double helix wraps around to form chromosomes. Methylation of histones can either increase or decrea ...
marks at the sites it binds, and this methyltransferase activity is essential for its role in DSB positioning. Following their formation, DSB sites are processed by resection, resulting in single-stranded DNA (ssDNA) that becomes decorated with DMC1. From mid-zygotene to early pachytene, as part of the recombinational repair process, DMC1 dissociates from the ssDNA and counts decrease until all breaks (except those on the XY chromosomes) are repaired at late pachytene. Several other proteins are involved in this process, including ZCWPW1, the first protein directly positioned by PRDM9's dual histone marks. ZCWPW1 is important for homologous DSB repair, not positioning.
Models
 Two primary models for how homologous recombination repairs double-strand breaks in DNA are the double-strand break repair (DSBR) pathway (sometimes called the ''double Holliday junction model'') and the synthesis-dependent strand annealing (SDSA) pathway. The two pathways are similar in their first several steps. After a double-strand break occurs, the MRX complex (
Two primary models for how homologous recombination repairs double-strand breaks in DNA are the double-strand break repair (DSBR) pathway (sometimes called the ''double Holliday junction model'') and the synthesis-dependent strand annealing (SDSA) pathway. The two pathways are similar in their first several steps. After a double-strand break occurs, the MRX complex (MRN complex
The MRN complex (MRX complex in yeast) is a protein complex consisting of Mre11, Rad50 and Nbs1 (also known as Nibrin in humans and as Xrs2 in yeast). In eukaryotes, the MRN/X complex plays an important role in the initial processing of double-st ...
in humans) binds to DNA on either side of the break. Next a resection takes place, in which DNA around the 5' ends of the break is cut back. This happens in two distinct steps: first the MRX complex recruits the Sae2 protein, and these two proteins trim back the 5' ends on either side of the break to create short 3' overhangs of single-strand DNA; in the second step, 5'→3' resection is continued by the Sgs1 helicase and the Exo1 and Dna2 DNA2 may refer to:
* '' DNA²'', a Japanese science fiction manga series
* DNA2 (gene)
DNA2-like helicase is an enzyme that in humans is encoded by the ''DNA2'' gene. Dna2, a homolog of DNA2KL present in budding yeast, possesses both helicase and ...
nucleases. As a helicase, Sgs1 "unzips" the double-strand DNA, while the nuclease
A nuclease (also archaically known as nucleodepolymerase or polynucleotidase) is an enzyme capable of cleaving the phosphodiester bonds between nucleotides of nucleic acids. Nucleases variously effect single and double stranded breaks in their ta ...
activity of Exo1 and Dna2 allows them to cut the single-stranded DNA produced by Sgs1.
The RPA protein, which has high affinity for single-stranded DNA, then binds the 3' overhangs. With the help of several other proteins that mediate the process, the Rad51
DNA repair protein RAD51 homolog 1 is a protein encoded by the gene ''RAD51''. The enzyme encoded by this gene is a member of the RAD51 protein family which assists in repair of DNA double strand breaks. RAD51 family members are homologous to th ...
protein (and Dmc1, in meiosis) then forms a filament of nucleic acid and protein on the single strand of DNA coated with RPA. This nucleoprotein filament then begins searching for DNA sequences similar to that of the 3' overhang. After finding such a sequence, the single-stranded nucleoprotein filament moves into (invades) the similar or identical recipient DNA duplex in a process called strand invasion. In cells that divide through mitosis, the recipient DNA duplex is generally a sister chromatid, which is identical to the damaged DNA molecule and provides a template for repair. In meiosis, however, the recipient DNA tends to be from a similar but not necessarily identical homologous chromosome. A displacement loop ( D-loop) is formed during strand invasion between the invading 3' overhang strand and the homologous chromosome. After strand invasion, a DNA polymerase extends the end of the invading 3' strand by synthesizing new DNA. This changes the D-loop to a cross-shaped structure known as a Holliday junction
A Holliday junction is a branched nucleic acid structure that contains four double-stranded arms joined. These arms may adopt one of several conformations depending on buffer salt concentrations and the sequence of nucleobases closest to the ju ...
. Following this, more DNA synthesis occurs on the invading strand (i.e., one of the original 3' overhangs), effectively restoring the strand on the homologous chromosome that was displaced during strand invasion.
DSBR pathway
nicking endonuclease A nicking enzyme (or nicking endonuclease) is an enzyme that cuts one strand of a double-stranded DNA at a specific recognition nucleotide
Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric u ...
s, a type of restriction endonuclease which cuts only one DNA strand. The DSBR pathway commonly results in crossover, though it can sometimes result in non-crossover products; the ability of a broken DNA molecule to collect sequences from separated donor loci was shown in mitotic budding yeast using plasmids or endonuclease induction of chromosomal events. Because of this tendency for chromosomal crossover, the DSBR pathway is a likely model of how crossover homologous recombination occurs during meiosis.
Whether recombination in the DSBR pathway results in chromosomal crossover is determined by how the double Holliday junction is cut, or "resolved". Chromosomal crossover will occur if one Holliday junction is cut on the crossing strand and the other Holliday junction is cut on the non-crossing strand (in Figure 5, along the horizontal purple arrowheads at one Holliday junction and along the vertical orange arrowheads at the other). Alternatively, if the two Holliday junctions are cut on the crossing strands (along the horizontal purple arrowheads at both Holliday junctions in Figure 5), then chromosomes without crossover will be produced.
SDSA pathway
Homologous recombination via the SDSA pathway occurs in cells that divide through mitosis and meiosis and results in non-crossover products. In this model, the invading 3' strand is extended along the recipient DNA duplex by a DNA polymerase, and is released as the Holliday junction between the donor and recipient DNA molecules slides in a process called ''branch migration''. The newly synthesized 3' end of the invading strand is then able to anneal to the other 3' overhang in the damaged chromosome through complementary base pairing. After the strands anneal, a small flap of DNA can sometimes remain. Any such flaps are removed, and the SDSA pathway finishes with the resealing, also known as '' ligation'', of any remaining single-stranded gaps. During mitosis, the major homologous recombination pathway for repairing DNA double-strand breaks appears to be the SDSA pathway (rather than the DSBR pathway). The SDSA pathway produces non-crossover recombinants (Figure 5). During meiosis non-crossover recombinants also occur frequently and these appear to arise mainly by the SDSA pathway as well. Non-crossover recombination events occurring during meiosis likely reflect instances of repair of DNA double-strand damages or other types of DNA damages.SSA pathway
mutagen
In genetics, a mutagen is a physical or chemical agent that permanently changes nucleic acid, genetic material, usually DNA, in an organism and thus increases the frequency of mutations above the natural background level. As many mutations can ca ...
ic since it results in such deletions of genetic material.
BIR pathway
During DNA replication, double-strand breaks can sometimes be encountered at replication forks as DNA helicase unzips the template strand. These defects are repaired in the ''break-induced replication'' (BIR) pathway of homologous recombination. The precise molecular mechanisms of the BIR pathway remain unclear. Three proposed mechanisms have strand invasion as an initial step, but they differ in how they model the migration of the D-loop and later phases of recombination. The BIR pathway can also help to maintain the length of telomeres (regions of DNA at the end of eukaryotic chromosomes) in the absence of (or in cooperation with) telomerase. Without working copies of the enzyme telomerase, telomeres typically shorten with each cycle of mitosis, which eventually blocks cell division and leads tosenescence
Senescence () or biological aging is the gradual deterioration of functional characteristics in living organisms. The word ''senescence'' can refer to either cellular senescence or to senescence of the whole organism. Organismal senescence inv ...
. In budding yeast cells where telomerase has been inactivated through mutations, two types of "survivor" cells have been observed to avoid senescence longer than expected by elongating their telomeres through BIR pathways.
Maintaining telomere length is critical for cell immortalization, a key feature of cancer. Most cancers maintain telomeres by upregulating telomerase. However, in several types of human cancer, a BIR-like pathway helps to sustain some tumors by acting as an alternative mechanism of telomere maintenance. This fact has led scientists to investigate whether such recombination-based mechanisms of telomere maintenance could thwart anti-cancer drugs like telomerase inhibitors.
In bacteria
 Homologous recombination is a major DNA repair process in bacteria. It is also important for producing genetic diversity in bacterial populations, although the process differs substantially from
Homologous recombination is a major DNA repair process in bacteria. It is also important for producing genetic diversity in bacterial populations, although the process differs substantially from meiotic
Meiosis (; , since it is a reductional division) is a special type of cell division of germ cells in sexually-reproducing organisms that produces the gametes, such as sperm or egg cells. It involves two rounds of division that ultimately res ...
recombination, which repairs DNA damages and brings about diversity in eukaryotic genomes. Homologous recombination has been most studied and is best understood for '' Escherichia coli''. Double-strand DNA breaks in bacteria are repaired by the RecBCD pathway of homologous recombination. Breaks that occur on only one of the two DNA strands, known as single-strand gaps, are thought to be repaired by the RecF pathway. Both the RecBCD and RecF pathways include a series of reactions known as '' branch migration'', in which single DNA strands are exchanged between two intercrossed molecules of duplex DNA, and ''resolution'', in which those two intercrossed molecules of DNA are cut apart and restored to their normal double-stranded state.
RecBCD pathway
The RecBCD pathway is the main recombination pathway used in many bacteria to repair double-strand breaks in DNA, and the proteins are found in a broad array of bacteria. These double-strand breaks can be caused by UV light and otherradiation
In physics, radiation is the emission or transmission of energy in the form of waves or particles through space or through a material medium. This includes:
* ''electromagnetic radiation'', such as radio waves, microwaves, infrared, visi ...
, as well as chemical mutagen
In genetics, a mutagen is a physical or chemical agent that permanently changes nucleic acid, genetic material, usually DNA, in an organism and thus increases the frequency of mutations above the natural background level. As many mutations can ca ...
s. Double-strand breaks may also arise by DNA replication through a single-strand nick or gap. Such a situation causes what is known as a collapsed replication fork and is fixed by several pathways of homologous recombination including the RecBCD pathway.
In this pathway, a three-subunit enzyme complex called RecBCD initiates recombination by binding to a blunt or nearly blunt end of a break in double-strand DNA. After RecBCD binds the DNA end, the RecB and RecD subunit
Subunit may refer to:
*Subunit HIV vaccine, a class of HIV vaccine
*Protein subunit, a protein molecule that assembles with other protein molecules
*Monomer, a molecule that may bind chemically to other molecules to form a polymer
*Sub-subunit, a ...
s begin unzipping the DNA duplex through helicase activity. The RecB subunit also has a nuclease
A nuclease (also archaically known as nucleodepolymerase or polynucleotidase) is an enzyme capable of cleaving the phosphodiester bonds between nucleotides of nucleic acids. Nucleases variously effect single and double stranded breaks in their ta ...
domain, which cuts the single strand of DNA that emerges from the unzipping process. This unzipping continues until RecBCD encounters a specific nucleotide sequence (5'-GCTGGTGG-3') known as a Chi site.
Upon encountering a Chi site, the activity of the RecBCD enzyme changes drastically. DNA unwinding pauses for a few seconds and then resumes at roughly half the initial speed. This is likely because the slower RecB helicase unwinds the DNA after Chi, rather than the faster RecD helicase, which unwinds the DNA before Chi. Recognition of the Chi site also changes the RecBCD enzyme so that it cuts the DNA strand with Chi and begins loading multiple RecA proteins onto the single-stranded DNA with the newly generated 3' end. The resulting RecA-coated nucleoprotein filament then searches out similar sequences of DNA on a homologous chromosome. The search process induces stretching of the DNA duplex, which enhances homology recognition (a mechanism termed conformational proofreading). Upon finding such a sequence, the single-stranded nucleoprotein filament moves into the homologous recipient DNA duplex in a process called ''strand invasion''. The invading 3' overhang causes one of the strands of the recipient DNA duplex to be displaced, to form a D-loop. If the D-loop is cut, another swapping of strands forms a cross-shaped structure called a Holliday junction
A Holliday junction is a branched nucleic acid structure that contains four double-stranded arms joined. These arms may adopt one of several conformations depending on buffer salt concentrations and the sequence of nucleobases closest to the ju ...
. Resolution of the Holliday junction by some combination of RuvABC or RecG can produce two recombinant DNA molecules with reciprocal genetic types, if the two interacting DNA molecules differ genetically. Alternatively, the invading 3’ end near Chi can prime DNA synthesis and form a replication fork. This type of resolution produces only one type of recombinant (non-reciprocal).
RecF pathway
Bacteria appear to use the RecF pathway of homologous recombination to repair single-strand gaps in DNA. When the RecBCD pathway is inactivated by mutations and additional mutations inactivate the SbcCD and ExoI nucleases, the RecF pathway can also repair DNA double-strand breaks. In the RecF pathway the RecQ helicase unwinds the DNA and the RecJ nuclease degrades the strand with a 5' end, leaving the strand with the 3' end intact. RecA protein binds to this strand and is either aided by the RecF, RecO, and RecR proteins or stabilized by them. The RecA nucleoprotein filament then searches for a homologous DNA and exchanges places with the identical or nearly identical strand in the homologous DNA. Although the proteins and specific mechanisms involved in their initial phases differ, the two pathways are similar in that they both require single-stranded DNA with a 3' end and the RecA protein for strand invasion. The pathways are also similar in their phases of '' branch migration'', in which the Holliday junction slides in one direction, and ''resolution'', in which the Holliday junctions are cleaved apart by enzymes. The alternative, non-reciprocal type of resolution may also occur by either pathway.Branch migration
Immediately after strand invasion, the Holliday junction moves along the linked DNA during the branch migration process. It is in this movement of the Holliday junction thatbase pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
s between the two homologous DNA duplexes are exchanged. To catalyze branch migration, the RuvA protein first recognizes and binds to the Holliday junction and recruits the RuvB protein to form the RuvAB complex. Two sets of the RuvB protein, which each form a ring-shaped ATPase
ATPases (, Adenosine 5'-TriPhosphatase, adenylpyrophosphatase, ATP monophosphatase, triphosphatase, SV40 T-antigen, ATP hydrolase, complex V (mitochondrial electron transport), (Ca2+ + Mg2+)-ATPase, HCO3−-ATPase, adenosine triphosphatase) are ...
, are loaded onto opposite sides of the Holliday junction, where they act as twin pumps that provide the force for branch migration. Between those two rings of RuvB, two sets of the RuvA protein assemble in the center of the Holliday junction such that the DNA at the junction is sandwiched between each set of RuvA. The strands of both DNA duplexes—the "donor" and the "recipient" duplexes—are unwound on the surface of RuvA as they are guided by the protein from one duplex to the other.
Resolution
In the resolution phase of recombination, any Holliday junctions formed by the strand invasion process are cut, thereby restoring two separate DNA molecules. This cleavage is done by RuvAB complex interacting with RuvC, which together form the RuvABC complex. RuvC is anendonuclease
Endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (without regard to sequence), while many, typically called restriction endonucleases ...
that cuts the degenerate
Degeneracy, degenerate, or degeneration may refer to:
Arts and entertainment
* ''Degenerate'' (album), a 2010 album by the British band Trigger the Bloodshed
* Degenerate art, a term adopted in the 1920s by the Nazi Party in Germany to descr ...
sequence 5'-(A/T)TT(G/C)-3'. The sequence is found frequently in DNA, about once every 64 nucleotides. Before cutting, RuvC likely gains access to the Holliday junction by displacing one of the two RuvA tetramers covering the DNA there. Recombination results in either "splice" or "patch" products, depending on how RuvC cleaves the Holliday junction. Splice products are crossover products, in which there is a rearrangement of genetic material around the site of recombination. Patch products, on the other hand, are non-crossover products in which there is no such rearrangement and there is only a "patch" of hybrid DNA in the recombination product.
Facilitating genetic transfer
Homologous recombination is an important method of integrating donor DNA into a recipient organism's genome in horizontal gene transfer, the process by which an organism incorporates foreign DNA from another organism without being the offspring of that organism. Homologous recombination requires incoming DNA to be highly similar to the recipient genome, and so horizontal gene transfer is usually limited to similar bacteria. Studies in several species of bacteria have established that there is a log-linear decrease in recombination frequency with increasing difference in sequence between host and recipient DNA. Inbacterial conjugation
Bacterial conjugation is the transfer of genetic material between bacterial cells by direct cell-to-cell contact or by a bridge-like connection between two cells. This takes place through a pilus. It is a parasexual mode of reproduction in bacteri ...
, where DNA is transferred between bacteria through direct cell-to-cell contact, homologous recombination helps integrate foreign DNA into the host genome via the RecBCD pathway. The RecBCD enzyme promotes recombination after DNA is converted from single-strand DNA–in which form it originally enters the bacterium–to double-strand DNA during replication. The RecBCD pathway is also essential for the final phase of transduction, a type of horizontal gene transfer in which DNA is transferred from one bacterium to another by a virus. Foreign, bacterial DNA is sometimes misincorporated in the capsid head of bacteriophage
A bacteriophage (), also known informally as a ''phage'' (), is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν ('), meaning "to devour". Bacteri ...
virus particles as DNA is packaged into new bacteriophages during viral replication. When these new bacteriophages infect other bacteria, DNA from the previous host bacterium is injected into the new bacterial host as double-strand DNA. The RecBCD enzyme then incorporates this double-strand DNA into the genome of the new bacterial host.
Bacterial transformation
Natural bacterial transformation involves the transfer of DNA from a donor bacterium to a recipient bacterium, where both donor and recipient are ordinarily of the same species. Transformation, unlike bacterial conjugation and transduction, depends on numerous bacterial gene products that specifically interact to perform this process. Thus transformation is clearly a bacterialadaptation
In biology, adaptation has three related meanings. Firstly, it is the dynamic evolutionary process of natural selection that fits organisms to their environment, enhancing their evolutionary fitness. Secondly, it is a state reached by the po ...
for DNA transfer. In order for a bacterium to bind, take up and integrate donor DNA into its resident chromosome by homologous recombination, it must first enter a special physiological state termed competence
Competence may refer to:
*Competence (geology), the resistance of a rock against deformation or plastic flow.
*Competence (human resources), a standardized requirement for an individual to properly perform a specific job
*Competence (law), the me ...
. The '' RecA''/ ''Rad51''/ ''DMC1'' gene family plays a central role in homologous recombination during bacterial transformation as it does during eukaryotic meiosis and mitosis. For instance, the RecA protein is essential for transformation in ''Bacillus subtilis'' and ''Streptococcus pneumoniae'', and expression of the RecA gene is induced during the development of competence for transformation in these organisms.
As part of the transformation process, the RecA protein interacts with entering single-stranded DNA (ssDNA) to form RecA/ssDNA nucleofilaments that scan the resident chromosome for regions of homology and bring the entering ssDNA to the corresponding region, where strand exchange and homologous recombination occur. Thus the process of homologous recombination during bacterial transformation has fundamental similarities to homologous recombination during meiosis.
In viruses
Homologous recombination occurs in several groups of viruses. In DNA viruses such as herpesvirus, recombination occurs through a break-and-rejoin mechanism like in bacteria and eukaryotes. There is also evidence for recombination in someRNA virus
An RNA virus is a virusother than a retrovirusthat has ribonucleic acid (RNA) as its genetic material. The nucleic acid is usually single-stranded RNA ( ssRNA) but it may be double-stranded (dsRNA). Notable human diseases caused by RNA viruses ...
es, specifically positive-sense ssRNA viruses like retrovirus
A retrovirus is a type of virus that inserts a DNA copy of its RNA genome into the DNA of a host cell that it invades, thus changing the genome of that cell. Once inside the host cell's cytoplasm, the virus uses its own reverse transcriptase ...
es, picornaviruses, and coronavirus
Coronaviruses are a group of related RNA viruses that cause diseases in mammals and birds. In humans and birds, they cause respiratory tract infections that can range from mild to lethal. Mild illnesses in humans include some cases of the com ...
es. There is controversy over whether homologous recombination occurs in negative-sense ssRNA viruses like influenza
Influenza, commonly known as "the flu", is an infectious disease caused by influenza viruses. Symptoms range from mild to severe and often include fever, runny nose, sore throat, muscle pain, headache, coughing, and fatigue. These symptoms ...
.
In RNA viruses, homologous recombination can be either precise or imprecise. In the precise type of RNA-RNA recombination, there is no difference between the two parental RNA sequences and the resulting crossover RNA region. Because of this, it is often difficult to determine the location of crossover events between two recombining RNA sequences. In imprecise RNA homologous recombination, the crossover region has some difference with the parental RNA sequences – caused by either addition, deletion, or other modification of nucleotides. The level of precision in crossover is controlled by the sequence context of the two recombining strands of RNA: sequences rich in adenine and uracil decrease crossover precision.
Homologous recombination is important in facilitating viral evolution. For example, if the genomes of two viruses with different disadvantageous mutations undergo recombination, then they may be able to regenerate a fully functional genome. Alternatively, if two similar viruses have infected the same host cell, homologous recombination can allow those two viruses to swap genes and thereby evolve more potent variations of themselves.
Homologous recombination is the proposed mechanism whereby the DNA virus '' human herpesvirus-6'' integrates into human telomeres.
When two or more viruses, each containing lethal genomic damage, infect the same host cell, the virus genomes can often pair with each other and undergo homologous recombinational repair to produce viable progeny. This process, known as multiplicity reactivation, has been studied in several bacteriophage
A bacteriophage (), also known informally as a ''phage'' (), is a duplodnaviria virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν ('), meaning "to devour". Bacteri ...
s, including phage T4
Escherichia virus T4 is a species of bacteriophages that infect ''Escherichia coli'' bacteria. It is a double-stranded DNA virus in the subfamily ''Tevenvirinae'' from the family Myoviridae. T4 is capable of undergoing only a lytic lifecycle ...
. Enzymes employed in recombinational repair in phage T4 are functionally homologous to enzymes employed in bacterial and eukaryotic recombinational repair. In particular, with regard to a gene necessary for the strand exchange reaction, a key step in homologous recombinational repair, there is functional homology from viruses to humans (i. e. ''uvsX'' in phage T4; '' recA'' in E. coli and other bacteria, and ''rad51
DNA repair protein RAD51 homolog 1 is a protein encoded by the gene ''RAD51''. The enzyme encoded by this gene is a member of the RAD51 protein family which assists in repair of DNA double strand breaks. RAD51 family members are homologous to th ...
'' and '' dmc1'' in yeast and other eukaryotes, including humans). Multiplicity reactivation has also been demonstrated in numerous pathogenic viruses.
Coronavirus
Coronaviruses are capable ofgenetic recombination
Genetic recombination (also known as genetic reshuffling) is the exchange of genetic material between different organisms which leads to production of offspring with combinations of traits that differ from those found in either parent. In eukaryo ...
when at least two viral genomes are present in the same infected cell. RNA recombination appears to be a major driving force in determining (1) genetic variability within a CoV species, (2) the capability of a CoV species to jump from one host to another, and (3) infrequently, the emergence of novel CoVs. The mechanism of recombination in CoVs likely involves template switching during genome replication. Recombination in RNA viruses appears to be an adaptation for coping with genome damage.
The pandemic SARS-CoV-2's entire receptor binding motif appears to have been introduced through recombination from coronavirus
Coronaviruses are a group of related RNA viruses that cause diseases in mammals and birds. In humans and birds, they cause respiratory tract infections that can range from mild to lethal. Mild illnesses in humans include some cases of the com ...
es of pangolin
Pangolins, sometimes known as scaly anteaters, are mammals of the order Pholidota (, from Ancient Greek ϕολιδωτός – "clad in scales"). The one extant family, the Manidae, has three genera: '' Manis'', '' Phataginus'', and '' Smut ...
s. Such a recombination event may have been a critical step in the evolution of SARS-CoV-2's capability to infect humans. Recombination events are likely key steps in the evolutionary process that leads to the emergence of new human coronaviruses.
During COVID-19 pandemic in 2020, many genomic sequences of Australian SARS‐CoV‐2 isolates have deletions or mutations (29742G>A or 29742G>U; "G19A" or "G19U")in the Coronavirus 3′ stem-loop II-like motif (s2m)
The Coronavirus 3′ stem-loop II-like motif (also known as s2m) is a secondary structure motif identified in the 3′ UTR, 3′ untranslated region (3′UTR) of astrovirus, coronavirus and equine rhinovirus genomes. Its function is unknown, but ...
, an RNA motif in 3' untranslated region of viral genome, suggesting that RNA recombination events may have occurred in s2m of SARS-CoV-2. Based on computational analysis of 1319 Australia SARS‐CoV‐2 sequences using Recco algorithm (https://recco.bioinf.mpi-inf.mpg.de/), 29742G("G19"), 29744G("G21"), and 29751G("G28") were predicted as recombination hotspots.  The SARS-CoV-2 outbreak in Diamond Princess cruise most likely originated from either a single person infected with a virus variant identical to the Wuhan WIV04 isolates, or simultaneously with another primary case infected with a virus containing the 11083G > T mutation. Linkage disequilibrium analysis confirmed that
The SARS-CoV-2 outbreak in Diamond Princess cruise most likely originated from either a single person infected with a virus variant identical to the Wuhan WIV04 isolates, or simultaneously with another primary case infected with a virus containing the 11083G > T mutation. Linkage disequilibrium analysis confirmed that RNA recombination
Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and deoxyribonucleic acid ( DNA) are nucleic acids. Along with lipids, proteins, and carbohydra ...
with the 11083G > T mutation also contributed to the increase of mutations among the viral progeny. The findings indicate that the 11083G > T mutation of SARS-CoV-2 spread during shipboard quarantine and arose through de novo RNA recombination
Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and deoxyribonucleic acid ( DNA) are nucleic acids. Along with lipids, proteins, and carbohydra ...
under positive selection pressure. In addition, in three patients in this cruise, two mutations 29736G > T and 29751G > T ("G13" and "G28") were also located in Coronavirus 3′ stem-loop II-like motif (s2m)
The Coronavirus 3′ stem-loop II-like motif (also known as s2m) is a secondary structure motif identified in the 3′ UTR, 3′ untranslated region (3′UTR) of astrovirus, coronavirus and equine rhinovirus genomes. Its function is unknown, but ...
, as "G28" was predicted as recombination hotspots in Australian SARS-CoV-2 mutants. Although s2m is considered an RNA motif highly conserved among many coronavirus species, this result also suggests that s2m of SARS-CoV-2 is rather a RNA recombination
Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and deoxyribonucleic acid ( DNA) are nucleic acids. Along with lipids, proteins, and carbohydra ...
/mutation hotspot.
Effects of dysfunction
 Without proper homologous recombination, chromosomes often incorrectly align for the first phase of cell division in meiosis. This causes chromosomes to fail to properly segregate in a process called nondisjunction. In turn, nondisjunction can cause
Without proper homologous recombination, chromosomes often incorrectly align for the first phase of cell division in meiosis. This causes chromosomes to fail to properly segregate in a process called nondisjunction. In turn, nondisjunction can cause sperm
Sperm is the male reproductive cell, or gamete, in anisogamous forms of sexual reproduction (forms in which there is a larger, female reproductive cell and a smaller, male one). Animals produce motile sperm with a tail known as a flagellum, whi ...
and ova to have too few or too many chromosomes. Down's syndrome, which is caused by an extra copy of chromosome 21, is one of many abnormalities that result from such a failure of homologous recombination in meiosis.
Deficiencies in homologous recombination have been strongly linked to cancer formation in humans. For example, each of the cancer-related diseases Bloom syndrome, Werner syndrome and Rothmund–Thomson syndrome are caused by malfunctioning copies of RecQ helicase genes involved in the regulation of homologous recombination: '' BLM'', '' WRN'' and '' RECQL4'', respectively. In the cells of Bloom's syndrome patients, who lack a working copy of the BLM protein, there is an elevated rate of homologous recombination. Experiments in mice deficient in BLM have suggested that the mutation gives rise to cancer through a loss of heterozygosity caused by increased homologous recombination. A loss in heterozygosity refers to the loss of one of two versions—or alleles—of a gene. If one of the lost alleles helps to suppress tumors, like the gene for the retinoblastoma protein for example, then the loss of heterozygosity can lead to cancer.
Decreased rates of homologous recombination cause inefficient DNA repair, which can also lead to cancer. This is the case with BRCA1 and BRCA2
''BRCA2'' and BRCA2 () are a human gene and its protein product, respectively. The official symbol (BRCA2, italic for the gene, nonitalic for the protein) and the official name (originally breast cancer 2; currently BRCA2, DNA repair associated) ...
, two similar tumor suppressor genes whose malfunctioning has been linked with considerably increased risk for breast and ovarian cancer. Cells missing BRCA1 and BRCA2 have a decreased rate of homologous recombination and increased sensitivity to ionizing radiation
Ionizing radiation (or ionising radiation), including nuclear radiation, consists of subatomic particles or electromagnetic waves that have sufficient energy to ionize atoms or molecules by detaching electrons from them. Some particles can travel ...
, suggesting that decreased homologous recombination leads to increased susceptibility to cancer. Because the only known function of BRCA2 is to help initiate homologous recombination, researchers have speculated that more detailed knowledge of BRCA2's role in homologous recombination may be the key to understanding the causes of breast and ovarian cancer.
Tumours with a homologous recombination deficiency (including BRCA defects) are described as HRD-positive.
Evolutionary conservation
Rad51
DNA repair protein RAD51 homolog 1 is a protein encoded by the gene ''RAD51''. The enzyme encoded by this gene is a member of the RAD51 protein family which assists in repair of DNA double strand breaks. RAD51 family members are homologous to th ...
and DMC1 in eukaryotes, RadA in archaea
Archaea ( ; singular archaeon ) is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria (in the Archaebac ...
, and UvsX in T4 phage
Escherichia virus T4 is a species of bacteriophages that infect ''Escherichia coli'' bacteria. It is a double-stranded DNA virus in the subfamily '' Tevenvirinae'' from the family Myoviridae. T4 is capable of undergoing only a lytic lifecycle ...
.
Related single stranded binding proteins that are important for homologous recombination, and many other processes, are also found in all domains of life.
Rad54, Mre11, Rad50, and a number of other proteins are also found in both archaea and eukaryotes.
The RecA recombinase family
The proteins of the RecA recombinase family of proteins are thought to be descended from a common ancestral recombinase. The RecA recombinase family contains RecA protein from bacteria, theRad51
DNA repair protein RAD51 homolog 1 is a protein encoded by the gene ''RAD51''. The enzyme encoded by this gene is a member of the RAD51 protein family which assists in repair of DNA double strand breaks. RAD51 family members are homologous to th ...
and Dmc1 proteins from eukaryotes, and RadA from archaea
Archaea ( ; singular archaeon ) is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria (in the Archaebac ...
, and the recombinase paralog proteins. Studies modeling the evolutionary relationships between the Rad51, Dmc1 and RadA proteins indicate that they are monophyletic
In cladistics for a group of organisms, monophyly is the condition of being a clade—that is, a group of taxa composed only of a common ancestor (or more precisely an ancestral population) and all of its lineal descendants. Monophyletic gro ...
, or that they share a common molecular ancestor. Within this protein family, Rad51 and Dmc1 are grouped together in a separate clade
A clade (), also known as a monophyletic group or natural group, is a group of organisms that are monophyletic – that is, composed of a common ancestor and all its lineal descendants – on a phylogenetic tree. Rather than the English term, ...
from RadA. One of the reasons for grouping these three proteins together is that they all possess a modified helix-turn-helix motif, which helps the proteins bind to DNA, toward their N-terminal ends. An ancient gene duplication
Gene duplication (or chromosomal duplication or gene amplification) is a major mechanism through which new genetic material is generated during molecular evolution. It can be defined as any duplication of a region of DNA that contains a gene. ...
event of a eukaryotic RecA gene and subsequent mutation has been proposed as a likely origin of the modern RAD51 and DMC1 genes.
The proteins generally share a long conserved region known as the RecA/Rad51 domain. Within this protein domain are two sequence motif
In biology, a sequence motif is a nucleotide or amino-acid sequence pattern that is widespread and usually assumed to be related to biological function of the macromolecule. For example, an ''N''-glycosylation site motif can be defined as ''As ...
s, Walker A motif and Walker B motif. The Walker A and B motifs allow members of the RecA/Rad51 protein family to engage in ATP binding and ATP hydrolysis.
Meiosis-specific proteins
The discovery of Dmc1 in several species of ''Giardia
''Giardia'' ( or ) is a genus of anaerobic flagellated protozoan parasites of the phylum Metamonada that colonise and reproduce in the small intestines of several vertebrates, causing the disease giardiasis. Their life cycle alternates between ...
'', one of the earliest protists
A protist () is any eukaryotic organism (that is, an organism whose cells contain a cell nucleus) that is not an animal, plant, or fungus. While it is likely that protists share a common ancestor (the last eukaryotic common ancestor), the excl ...
to diverge as a eukaryote, suggests that meiotic homologous recombination—and thus meiosis itself—emerged very early in eukaryotic evolution. In addition to research on Dmc1, studies on the Spo11
Spo11 is a protein that in humans is encoded by the ''SPO11'' gene. Spo11, in a complex with mTopVIB, creates double strand breaks to initiate meiotic recombination. Its active site contains a tyrosine which ligates and dissociates with DNA to pr ...
protein have provided information on the origins of meiotic recombination. Spo11, a type II topoisomerase, can initiate homologous recombination in meiosis by making targeted double-strand breaks in DNA. Phylogenetic tree
A phylogenetic tree (also phylogeny or evolutionary tree Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA.) is a branching diagram or a tree showing the evolutionary relationships among various biological spec ...
s based on the sequence of genes similar to SPO11 in animals, fungi, plants, protists and archaea have led scientists to believe that the version Spo11 currently in eukaryotes emerged in the last common ancestor of eukaryotes and archaea.
Technological applications
Gene targeting
 Many methods for introducing DNA sequences into organisms to create recombinant DNA and
Many methods for introducing DNA sequences into organisms to create recombinant DNA and genetically modified organism
A genetically modified organism (GMO) is any organism whose genetic material has been altered using genetic engineering techniques. The exact definition of a genetically modified organism and what constitutes genetic engineering varies, with ...
s use the process of homologous recombination. Also called gene targeting, the method is especially common in yeast and mouse
A mouse ( : mice) is a small rodent. Characteristically, mice are known to have a pointed snout, small rounded ears, a body-length scaly tail, and a high breeding rate. The best known mouse species is the common house mouse (''Mus musculus' ...
genetics. The gene targeting method in knockout mice
A knockout mouse, or knock-out mouse, is a genetically modified mouse (''Mus musculus'') in which researchers have inactivated, or "knocked out", an existing gene by replacing it or disrupting it with an artificial piece of DNA. They are importan ...
uses mouse embryonic stem cells to deliver artificial genetic material (mostly of therapeutic interest), which represses the target gene of the mouse by the principle of homologous recombination. The mouse thereby acts as a working model to understand the effects of a specific mammalian gene. In recognition of their discovery of how homologous recombination can be used to introduce genetic modifications in mice through embryonic stem cells, Mario Capecchi, Martin Evans and Oliver Smithies were awarded the 2007 Nobel Prize for Physiology or Medicine.
Advances in gene targeting technologies which hijack the homologous recombination mechanics of cells are now leading to the development of a new wave of more accurate, isogenic human disease models. These engineered human cell models are thought to more accurately reflect the genetics of human diseases than their mouse model predecessors. This is largely because mutations of interest are introduced into endogenous genes, just as they occur in the real patients, and because they are based on human genomes rather than rat genomes. Furthermore, certain technologies enable the knock-in of a particular mutation rather than just knock-outs associated with older gene targeting technologies.
Protein engineering
Protein engineering
Protein engineering is the process of developing useful or valuable proteins. It is a young discipline, with much research taking place into the understanding of protein folding and recognition for protein design principles. It has been used to imp ...
with homologous recombination develops chimeric proteins by swapping fragments between two parental proteins. These techniques exploit the fact that recombination can introduce a high degree of sequence diversity while preserving a protein's ability to fold into its tertiary structure, or three-dimensional shape. This stands in contrast to other protein engineering techniques, like random point mutagenesis, in which the probability of maintaining protein function declines exponentially with increasing amino acid substitutions. The chimeras produced by recombination techniques are able to maintain their ability to fold because their swapped parental fragments are structurally and evolutionarily conserved. These recombinable "building blocks" preserve structurally important interactions like points of physical contact between different amino acids in the protein's structure. Computational methods like SCHEMA and statistical coupling analysis
Statistical coupling analysis or SCA is a technique used in bioinformatics to measure covariation between pairs of amino acids in a protein multiple sequence alignment (MSA). More specifically, it quantifies how much the amino acid distribution ...
can be used to identify structural subunits suitable for recombination.
Techniques that rely on homologous recombination have been used to engineer new proteins. In a study published in 2007, researchers were able to create chimeras of two enzymes involved in the biosynthesis of isoprenoid
The terpenoids, also known as isoprenoids, are a class of naturally occurring organic chemicals derived from the 5-carbon compound isoprene and its derivatives called terpenes, diterpenes, etc. While sometimes used interchangeably with "terpenes", ...
s, a diverse class of compounds including hormones, visual pigments and certain pheromones. The chimeric proteins acquired an ability to catalyze an essential reaction in isoprenoid biosynthesis—one of the most diverse pathways of biosynthesis
Biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined to form macromolecules. ...
found in nature—that was absent in the parent proteins. Protein engineering through recombination has also produced chimeric enzymes with new function in members of a group of proteins known as the cytochrome P450
Cytochromes P450 (CYPs) are a Protein superfamily, superfamily of enzymes containing heme as a cofactor (biochemistry), cofactor that functions as monooxygenases. In mammals, these proteins oxidize steroids, fatty acids, and xenobiotics, and are ...
family, which in humans is involved in detoxifying
Detoxification or detoxication (detox for short) is the physiological or medicinal removal of toxic substances from a living organism, including the human body, which is mainly carried out by the liver. Additionally, it can refer to the period of ...
foreign compounds like drugs, food additives and preservatives.
Cancer therapy
Homologous recombination proficient (HRP) cancer cells are able to repair the DNA damage, which is caused by chemotherapy such as cisplatin. Thus, HRP cancers are difficult to treat. Studies suggest that homologous recombination can be targeted via c-Abl inhibition. Cancer cells with BRCA mutations have deficiencies in homologous recombination, and drugs to exploit those deficiencies have been developed and used successfully in clinical trials. Olaparib, a PARP1 inhibitor, shrunk or stopped the growth of tumors from breast, ovarian andprostate cancer
Prostate cancer is cancer of the prostate. Prostate cancer is the second most common cancerous tumor worldwide and is the fifth leading cause of cancer-related mortality among men. The prostate is a gland in the male reproductive system that sur ...
s caused by mutations in the BRCA1 or BRCA2
''BRCA2'' and BRCA2 () are a human gene and its protein product, respectively. The official symbol (BRCA2, italic for the gene, nonitalic for the protein) and the official name (originally breast cancer 2; currently BRCA2, DNA repair associated) ...
genes, which are necessary for HR. When BRCA1 or BRCA2 is absent, other types of DNA repair mechanisms must compensate for the deficiency of HR, such as base-excision repair (BER) for stalled replication forks or non-homologous end joining (NHEJ) for double strand breaks. By inhibiting BER in an HR-deficient cell, olaparib applies the concept of synthetic lethality to specifically target cancer cells. While PARP1 inhibitors represent a novel approach to cancer therapy, researchers have cautioned that they may prove insufficient for treating late-stage metastatic cancers. Cancer cells can become resistant to a PARP1 inhibitor if they undergo deletions of mutations in BRCA2, undermining the drug's synthetic lethality by restoring cancer cells' ability to repair DNA by HR.
See also
*Chromosomal crossover
Chromosomal crossover, or crossing over, is the exchange of genetic material during sexual reproduction between two homologous chromosomes' non-sister chromatids that results in recombinant chromosomes. It is one of the final phases of geneti ...
* Homology directed repair
References
External links
Animations – homologous recombination
Animations showing several models of homologous recombination
Animation of the bacterial RecBCD pathway of homologous recombination {{DEFAULTSORT:Homologous Recombination DNA repair Modification of genetic information Telomeres