Haplogroup B-M108 on:
[Wikipedia]
[Google]
[Amazon]
A haplotype is a group of  Each haplogroup originates from, and remains part of, a preceding single haplogroup (or
Each haplogroup originates from, and remains part of, a preceding single haplogroup (or
 Mitochondria are small organelles that lie in the
Mitochondria are small organelles that lie in the
 Human
Human
 Haplogroups can be used to define genetic populations and are often geographically oriented. For example, the following are common divisions for mtDNA haplogroups:
* African: L0, L1, L2, L3, L4, L5, L6
* West Eurasian: H, T, U, V, X, K, I, J, W (all listed West Eurasian haplogroups are derived from macro-haplogroup N)
* East Eurasian: A, B, C, D, E, F, G, Y, Z (note: C, D, E, G, and Z belong to macro-haplogroup M)
* Native American: A, B, C, D, X
*
Haplogroups can be used to define genetic populations and are often geographically oriented. For example, the following are common divisions for mtDNA haplogroups:
* African: L0, L1, L2, L3, L4, L5, L6
* West Eurasian: H, T, U, V, X, K, I, J, W (all listed West Eurasian haplogroups are derived from macro-haplogroup N)
* East Eurasian: A, B, C, D, E, F, G, Y, Z (note: C, D, E, G, and Z belong to macro-haplogroup M)
* Native American: A, B, C, D, X
*
The Genographic Project
Y Chromosome Consortium
PhyloTree's Y-tree
A minimal reference phylogeny for the human Y-chromosome
* The Y Chromosome Consortium (2002)
A Nomenclature System for the Tree of Human Y-Chromosomal Binary Haplogroups
Genome Research, Vol. 12(2), 339–48, February 2002. (Detailed hierarchica
chart
has conversions from previous naming schemes) * Semino et al. (2000)
The Genetic Legacy of Paleolithic Homo sapiens sapiens in Extant Europeans
Science, Vol 290 (paper which introduced the "Eu" haplogroups).
Y-DNA Ethnographic and Genographic Atlas and Open-Source Data Compilation
PhyloD3 – D3.js-based phylogenetic tree based on PhyloTree
MitoTool – a web server for the analysis and retrieval of human mitochondrial DNA sequence variations
HaploGrep – automatic classification of mitochondrial DNA haplogroups based on PhyloTree
{{Webarchive, url=https://web.archive.org/web/20160612075145/http://haplogrep.uibk.ac.at/ , date=2016-06-12
HaploFind – fast automatic haplogroup assignment pipeline for human mitochondrial DNA
graphical mtDNA haplogroup skeleton
The Making of the African mtDNA Landscape
Do the Four Clades of the mtDNA Haplogroup L2 Evolve at Different Rates?
Y-DNA Haplogroup Browser
DNA Human evolution Phylogenetics Population genetics Classical genetics
allele
An allele (, ; ; modern formation from Greek ἄλλος ''állos'', "other") is a variation of the same sequence of nucleotides at the same place on a long DNA molecule, as described in leading textbooks on genetics and evolution.
::"The chro ...
s in an organism that are inherited together from a single parent, and a haplogroup ( haploid from the el, ἁπλοῦς, ''haploûs'', "onefold, simple" and en, group) is a group of similar haplotypes that share a common ancestor with a single-nucleotide polymorphism
In genetics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently lar ...
mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA replication, DNA or viral repl ...
. More specifically, a haplogroup is a combination of allele
An allele (, ; ; modern formation from Greek ἄλλος ''állos'', "other") is a variation of the same sequence of nucleotides at the same place on a long DNA molecule, as described in leading textbooks on genetics and evolution.
::"The chro ...
s at different chromosomal regions that are closely linked and that tend to be inherited together. As a haplogroup consists of similar haplotypes, it is usually possible to predict a haplogroup from haplotypes. Haplogroups pertain to a single line of descent. As such, membership of a haplogroup, by any individual, relies on a relatively small proportion of the genetic material possessed by that individual.
 Each haplogroup originates from, and remains part of, a preceding single haplogroup (or
Each haplogroup originates from, and remains part of, a preceding single haplogroup (or paragroup
Paragroup is a term used in population genetics to describe lineages within a haplogroup that are not defined by any additional unique markers.
In human Y-chromosome DNA haplogroups, paragroups are typically represented by an asterisk (*) placed ...
). As such, any related group of haplogroups may be precisely modelled as a nested hierarchy
A hierarchy (from Greek: , from , 'president of sacred rites') is an arrangement of items (objects, names, values, categories, etc.) that are represented as being "above", "below", or "at the same level as" one another. Hierarchy is an important ...
, in which each set (haplogroup) is also a subset of a single broader set (as opposed, that is, to biparental models, such as human family trees).
Haplogroups are normally identified by an initial letter of the alphabet, and refinements consist of additional number and letter combinations, such as (for example) .
In human genetics
Human genetics is the study of inheritance as it occurs in human beings. Human genetics encompasses a variety of overlapping fields including: classical genetics, cytogenetics, molecular genetics, biochemical genetics, genomics, population gene ...
, the haplogroups most commonly studied are Y-chromosome (Y-DNA) haplogroups and mitochondrial DNA (mtDNA) haplogroups, each of which can be used to define genetic populations
Population genetics is a subfield of genetics that deals with genetic differences within and between populations, and is a part of evolutionary biology. Studies in this branch of biology examine such phenomena as adaptation, speciation, and popu ...
. Y-DNA is passed solely along the patrilineal
Patrilineality, also known as the male line, the spear side or agnatic kinship, is a common kinship system in which an individual's family membership derives from and is recorded through their father's lineage. It generally involves the inheritan ...
line, from father to son, while mtDNA is passed down the matrilineal
Matrilineality is the tracing of kinship through the female line. It may also correlate with a social system in which each person is identified with their matriline – their mother's lineage – and which can involve the inheritance ...
line, from mother to offspring of both sexes. Neither recombines, and thus Y-DNA and mtDNA change only by chance mutation at each generation with no intermixture between parents' genetic material.
Haplogroup formation
 Mitochondria are small organelles that lie in the
Mitochondria are small organelles that lie in the cytoplasm
In cell biology, the cytoplasm is all of the material within a eukaryotic cell, enclosed by the cell membrane, except for the cell nucleus. The material inside the nucleus and contained within the nuclear membrane is termed the nucleoplasm. ...
of eukaryotic cells
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bact ...
, such as those of humans. Their primary function is to provide energy to the cell. Mitochondria are thought to be reduced descendants of symbiotic bacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one Cell (biology), biological cell. They constitute a large domain (biology), domain of prokaryotic microorganisms. Typically a few micrometr ...
that were once free living. One indication that mitochondria were once free living is that each contains a circular DNA, called mitochondrial DNA (mtDNA), whose structure is more similar to bacteria than eukaryotic organisms (see endosymbiotic theory). The overwhelming majority of a human's DNA is contained in the chromosome
A chromosome is a long DNA molecule with part or all of the genetic material of an organism. In most chromosomes the very long thin DNA fibers are coated with packaging proteins; in eukaryotic cells the most important of these proteins are ...
s in the nucleus
Nucleus ( : nuclei) is a Latin word for the seed inside a fruit. It most often refers to:
*Atomic nucleus, the very dense central region of an atom
* Cell nucleus, a central organelle of a eukaryotic cell, containing most of the cell's DNA
Nucl ...
of the cell, but mtDNA is an exception.
An individual inherits their cytoplasm and the organelles contained by that cytoplasm exclusively from the maternal ovum (egg cell); sperm only pass on the chromosomal DNA, all paternal mitochondria are digested in the oocyte
An oocyte (, ), oöcyte, or ovocyte is a female gametocyte or germ cell involved in reproduction. In other words, it is an immature ovum, or egg cell. An oocyte is produced in a female fetus in the ovary during female gametogenesis. The femal ...
. When a mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA replication, DNA or viral repl ...
arises in a mtDNA molecule, the mutation is therefore passed in a direct female line of descent. Mutations are copying mistakes in the DNA sequence. Single mistakes are called single nucleotide polymorphisms (SNPs).
Human Y chromosome
The Y chromosome is one of two sex chromosomes (allosomes) in therian mammals, including humans, and many other animals. The other is the X chromosome. Y is normally the sex-determining chromosome in many species, since it is the presence or abse ...
s are male-specific sex chromosomes
A sex chromosome (also referred to as an allosome, heterotypical chromosome, gonosome, heterochromosome, or idiochromosome) is a chromosome that differs from an ordinary autosome in form, size, and behavior. The human sex chromosomes, a typical ...
; nearly all humans that possess a Y chromosome will be morphologically male. Although Y chromosomes are situated in the cell nucleus and paired with X chromosome
The X chromosome is one of the two sex-determining chromosomes (allosomes) in many organisms, including mammals (the other is the Y chromosome), and is found in both males and females. It is a part of the XY sex-determination system and XO sex ...
s, they only recombine with the X chromosome at the ends of the Y chromosome; the remaining 95% of the Y chromosome does not recombine. Therefore, the Y chromosome and any mutations that arise in it are passed on from father to son in a direct male line of descent. This means the Y chromosome and mtDNA share specific properties.
Other chromosomes, autosomes and X chromosomes
The X chromosome is one of the two sex-determining chromosomes (allosomes) in many organisms, including mammals (the other is the Y chromosome), and is found in both males and females. It is a part of the XY sex-determination system and XO sex-d ...
in women, share their genetic material (called crossing over leading to recombination) during meiosis
Meiosis (; , since it is a reductional division) is a special type of cell division of germ cells in sexually-reproducing organisms that produces the gametes, such as sperm or egg cells. It involves two rounds of division that ultimately r ...
(a special type of cell division
Cell division is the process by which a parent cell divides into two daughter cells. Cell division usually occurs as part of a larger cell cycle in which the cell grows and replicates its chromosome(s) before dividing. In eukaryotes, there ar ...
that occurs for the purposes of sexual reproduction
Sexual reproduction is a type of reproduction that involves a complex life cycle in which a gamete ( haploid reproductive cells, such as a sperm or egg cell) with a single set of chromosomes combines with another gamete to produce a zygote th ...
). Effectively this means that the genetic material from these chromosomes gets mixed up in every generation, and so any new mutations are passed down randomly from parents to offspring.
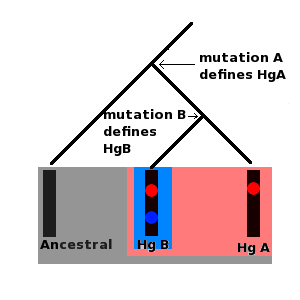
The special feature that both Y chromosomes and mtDNA display is that mutations can accrue along a certain segment of both molecules and these mutations remain fixed in place on the DNA. Furthermore, the historical sequence of these mutations can also be inferred. For example, if a set of ten Y chromosomes (derived from ten different men) contains a mutation, A, but only five of these chromosomes contain a second mutation, B, then it must be the case that mutation B occurred after mutation A.
Furthermore, all ten men who carry the chromosome with mutation A are the direct male line descendants of the same man who was the first person to carry this mutation. The first man to carry mutation B was also a direct male line descendant of this man, but is also the direct male line ancestor of all men carrying mutation B. Series of mutations such as this form molecular lineages. Furthermore, each mutation defines a set of specific Y chromosomes called a haplogroup.
All men carrying mutation A form a single haplogroup, and all men carrying mutation B are part of this haplogroup, but mutation B also defines a more recent haplogroup (which is a subgroup or subclade
In genetics, a subclade is a subgroup of a haplogroup.
Naming convention
Although human mitochondrial DNA (mtDNA) and Y chromosome DNA (Y-DNA) haplogroups and subclades are named in a similar manner, their names belong to completely separate sy ...
) of its own to which men carrying only mutation A do not belong. Both mtDNA and Y chromosomes are grouped into lineages and haplogroups; these are often presented as tree like diagrams.
Haplogroup population genetics
It is usually assumed that there is littlenatural selection
Natural selection is the differential survival and reproduction of individuals due to differences in phenotype. It is a key mechanism of evolution, the change in the heritable traits characteristic of a population over generations. Cha ...
for or against a particular haplotype mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA replication, DNA or viral repl ...
which has survived to the present day, so apart from mutation rate
In genetics, the mutation rate is the frequency of new mutations in a single gene or organism over time. Mutation rates are not constant and are not limited to a single type of mutation; there are many different types of mutations. Mutation rates ...
s (which may vary from one marker to another) the main driver of population genetics
Population genetics is a subfield of genetics that deals with genetic differences within and between populations, and is a part of evolutionary biology. Studies in this branch of biology examine such phenomena as Adaptation (biology), adaptation, ...
affecting the proportions of haplotypes in a population is genetic drift
Genetic drift, also known as allelic drift or the Wright effect, is the change in the frequency of an existing gene variant (allele) in a population due to random chance.
Genetic drift may cause gene variants to disappear completely and there ...
—random fluctuation caused by the sampling randomness of which members of the population happen to pass their DNA on to members of the next generation of the appropriate sex.
This causes the prevalence of a particular marker in a population to continue to fluctuate, until it either hits 100%, or falls out of the population entirely. In a large population with efficient mixing the rate of genetic drift for common alleles is very low; however, in a very small interbreeding population the proportions can change much more quickly. The marked geographical variations and concentrations of particular haplotypes and groups of haplotypes therefore witness the distinctive effects of repeated population bottlenecks or founder events followed by population separations and increases.
The lineages which can be traced back from the present will not reflect the full genetic variation of the older population: genetic drift means that some of the variants will have died out. The cost of full Y-DNA and mtDNA sequence tests has limited the availability of data; however, their cost has dropped dramatically in the last decade. Haplotype coalescence times and current geographical prevalences both carry considerable error uncertainties. This is especially troublesome for coalescence times, because most population geneticists still continue (albeit decreasing a little bit) to use the "Zhivotovski method", which is heavily criticised by DNA-genealogists for its falsehood. The eusocial wasp '' Angiopolybia pallens ''presents with 8 haplogroups depending on its location. This displays the idea of genetic drift.
Human Y-chromosome DNA haplogroups
HumanY chromosome
The Y chromosome is one of two sex chromosomes (allosomes) in therian mammals, including humans, and many other animals. The other is the X chromosome. Y is normally the sex-determining chromosome in many species, since it is the presence or abse ...
DNA (Y-DNA) haplogroups are named from A to T, and are further subdivided using numbers and lower case letters. Y chromosome haplogroup designations are established by the Y Chromosome Consortium.
Y-chromosomal Adam
In human genetics, the Y-chromosomal most recent common ancestor (Y-MRCA, informally known as Y-chromosomal Adam) is the patrilineal most recent common ancestor (MRCA) from whom all currently living humans are descended. He is the most recent mal ...
is the name given by researchers to the male who is the most recent common patrilineal (male-lineage) ancestor of all living humans.
Major Y-chromosome haplogroups, and their geographical regions of occurrence (prior to
the recent European colonization), include:
Groups without mutation M168
* Haplogroup A (M91) (Africa, especially the Khoisan andNilotes
The Nilotic peoples are people indigenous to the Nile Valley who speak Nilotic languages. They inhabit South Sudan, Sudan, Ethiopia, Uganda, Kenya, the Democratic Republic of the Congo, Rwanda, Burundi and Tanzania. Among these are the ...
)
* Haplogroup B (M60) (Africa, especially the Pygmies
In anthropology, pygmy peoples are ethnic groups whose average height is unusually short. The term pygmyism is used to describe the phenotype of endemic short stature (as opposed to disproportionate dwarfism occurring in isolated cases in a pop ...
and Hadzabe)
Groups with mutation M168
(mutation M168 occurred ~50,000 bp) * Haplogroup C (M130) (Oceania, North/Central/East Asia, North America and a minor presence in South America, Southeast Asia, South Asia, West Asia, and Europe) * YAP+ haplogroups ** Haplogroup DE (M1, M145, M203) *** Haplogroup D (CTS3946) (Tibet, Japan, the Andaman Islands, Central Asia, and a sporadic presence in Nigeria, Syria, and Saudi Arabia) *** Haplogroup E (M96) ****Haplogroup E1b1a
Haplogroup E-V38 is a human Y-chromosome DNA haplogroup. It is primarily distributed in Sub Saharan Africa. E-V38 has two basal branches, E-M329 (formerly E1b1c or E1b1*) and E-M2 (formerly E3a & E1b1a). The E-M329 subclade is today almost excl ...
(V38) West Africa and surrounding regions; formerly known as E3a
**** Haplogroup E1b1b (M215) Associated with the spread of Afroasiatic languages; now concentrated in North Africa and the Horn of Africa, as well as parts of the Middle East, the Mediterranean, and the Balkans; formerly known as E3b
Groups with mutation M89
(mutation M89 occurred ~45,000 bp) * Haplogroup F (M89) Oceania, Europe, Asia, North and South America ** Haplogroup FT (P14, M213) (China, Vietnam, Singapore) * Haplogroup G (M201) (present among many ethnic groups in Eurasia, usually at low frequency; most common in the Caucasus, theIranian plateau
The Iranian plateau or Persian plateau is a geological feature in Western Asia, Central Asia, and South Asia. It comprises part of the Eurasian Plate and is wedged between the Arabian Plate and the Indian Plate; situated between the Zagros ...
, and Anatolia; in Europe mainly in Greece, Italy, Iberia
The Iberian Peninsula (),
**
* Aragonese and Occitan: ''Peninsula Iberica''
**
**
* french: Péninsule Ibérique
* mwl, Península Eibérica
* eu, Iberiar penintsula also known as Iberia, is a peninsula in southwestern Europe, defi ...
, the Tyrol, Bohemia; rare in Northern Europe)
* Haplogroup H (L901/M2939)
**H1'3 (Z4221/M2826, Z13960)
***H1 (L902/M3061)
****H1a (M69/Page45) India, Sri Lanka, Nepal, Pakistan, Iran, Central Asia
****H1b (B108) Found in a Burmese individual in Myanmar
Myanmar, ; UK pronunciations: US pronunciations incl. . Note: Wikipedia's IPA conventions require indicating /r/ even in British English although only some British English speakers pronounce r at the end of syllables. As John Wells explai ...
.
***H3 (Z5857) India, Sri Lanka, Pakistan, Bahrain, Qatar
**H2 (P96) Formerly known as haplogroup F3. Found with low frequency in Europe and western Asia.
* Haplogroup IJK
Haplogroup IJK is a human Y-chromosome DNA haplogroup. IJK is a primary branch of the macrohaplogroup HIJK. Its direct descendants are haplogroup IJ and haplogroup K.
Distribution and structure
The basal paragroup HIJK* has been found in a Meso ...
(L15, L16)
Groups with mutations L15 & L16
*Haplogroup IJK
Haplogroup IJK is a human Y-chromosome DNA haplogroup. IJK is a primary branch of the macrohaplogroup HIJK. Its direct descendants are haplogroup IJ and haplogroup K.
Distribution and structure
The basal paragroup HIJK* has been found in a Meso ...
(L15, L16)
** Haplogroup IJ
Haplogroup IJ (M429/P125) is a human Y-chromosome DNA haplogroup, an immediate descendant of Haplogroup IJK (formerly known as Haplogroup F-L15). IJK is a branch of Haplogroup HIJK.
The immediate descendants of IJ are Haplogroup I and Haplogrou ...
(S2, S22)
*** Haplogroup I (M170, P19, M258) (widespread in Europe, found infrequently in parts of the Middle East, and virtually absent elsewhere)
**** Haplogroup I1
Haplogroup I-M253, also known as I1, is a Y chromosome haplogroup. The genetic markers confirmed as identifying I-M253 are the SNPs M253,M307.2/P203.2, M450/S109, P30, P40, L64, L75, L80, L81, L118, L121/S62, L123, L124/S64, L125/S65, L157.1, L1 ...
(M253, M307, P30, P40) (Northern Europe, dominant in Scandinavia)
**** Haplogroup I2 (S31) (Central and Southeast Europe, Sardinia, Balkans)
*** Haplogroup J (M304) (the Middle East, Turkey, Caucasus, Italy, Greece, the Balkans, North Africa)
**** Haplogroup J* (Mainly found in Socotra
Socotra or Soqotra (; ar, سُقُطْرَىٰ ; so, Suqadara) is an island of the Republic of Yemen in the Indian Ocean, under the ''de facto'' control of the UAE-backed Southern Transitional Council, a secessionist participant in Yemen’ ...
, with a few observations in Pakistan
Pakistan ( ur, ), officially the Islamic Republic of Pakistan ( ur, , label=none), is a country in South Asia. It is the world's fifth-most populous country, with a population of almost 243 million people, and has the world's second-lar ...
, Oman
Oman ( ; ar, عُمَان ' ), officially the Sultanate of Oman ( ar, سلْطنةُ عُمان ), is an Arabian country located in southwestern Asia. It is situated on the southeastern coast of the Arabian Peninsula, and spans the mouth of ...
, Greece
Greece,, or , romanized: ', officially the Hellenic Republic, is a country in Southeast Europe. It is situated on the southern tip of the Balkans, and is located at the crossroads of Europe, Asia, and Africa. Greece shares land borders ...
, the Czech Republic
The Czech Republic, or simply Czechia, is a landlocked country in Central Europe. Historically known as Bohemia, it is bordered by Austria to the south, Germany to the west, Poland to the northeast, and Slovakia to the southeast. The ...
, and among Turkic peoples
The Turkic peoples are a collection of diverse ethnic groups of West, Central, East, and North Asia as well as parts of Europe, who speak Turkic languages.. "Turkic peoples, any of various peoples whose members speak languages belonging t ...
)
**** Haplogroup J1 (M267) (Mostly associated with Semitic peoples in the Middle East but also found in; Mediterranean Europe, Ethiopia, North Africa, Iran, Pakistan, India and with Northeast Caucasian
The Northeast Caucasian languages, also called East Caucasian, Nakh-Daghestani or ''Vainakh-Daghestani'', is a family of languages spoken in the Russian republics of Dagestan, Chechnya and Ingushetia and in Northern Azerbaijan as well as ...
peoples in Dagestan; J1 with DYS388=13 is associated with eastern Anatolia)
**** Haplogroup J2
In human genetics, Haplogroup J-M172 or J2 is a Y-chromosome haplogroup which is a subclade (branch) of haplogroup J-M304. Haplogroup J-M172 is common in modern populations in Western Asia, Central Asia, South Asia, Europe, Northwestern Iran and ...
(M172) (Mainly found in West Asia, Central Asia, Southern Europe, and North Africa)
** Haplogroup K (M9, P128, P131, P132)
Groups with mutation M9
(mutation M9 occurred ~40,000 bp) * Haplogroup K **Haplogroup LT
Haplogroup LT or LT-L298/P326, also known as K1 (and until 2008 as Haplogroup K2), is a Y-chromosome DNA haplogroup. Its defining SNP mutations are L298 and P326.
No confirmed cases of the basal paragroup LT* have been identified among living ma ...
(L298/P326)
*** Haplogroup L (M11, M20, M22, M61, M185, M295) (South Asia, Central Asia, Southwestern Asia, the Mediterranean)
*** Haplogroup T (M70, M184/USP9Y+3178, M193, M272) (North Africa, Horn of Africa, Southwest Asia, the Mediterranean, South Asia); formerly known as Haplogroup K2
** Haplogroup K(xLT) (rs2033003/M526)
= Groups with mutation M526
= ** Haplogroup M (P256) (New Guinea, Melanesia, eastern Indonesia) **Haplogroup NO Haplogroup NO (M214/Page39; F176/M2314; CTS5858/M2325/F346; CTS11572), also known as NO-M214 and NO1, is a human Y-chromosome DNA haplogroup. NO is the sole confirmed subclade of Haplogroup K2a1 (K-M2313), which is the sole subclade of Haplogroup ...
(M214)
*** Haplogroup N (M231) (northernmost Eurasia)
*** Haplogroup O (M175) (East Asia, Southeast Asia, the South Pacific, South Asia, Central Asia)
**** Haplogroup O1 (F265)
***** Haplogroup O1a (M119)
***** Haplogroup O1b (P31, M268)
**** Haplogroup O2 (M122)
** Haplogroup P-M45
Haplogroup P1, also known as P-M45 and K2b2a, is a Y-chromosome DNA haplogroup in human genetics. Defined by the SNPs M45 and PF5962, P1 is a primary branch (subclade) of P (P-P295; K2b2).
The only primary subclades of P1 are Haplogroup Q (Q- ...
(M45) (M45 occurred ~35,000 bp)
*** Haplogroup Q-M242
Haplogroup Q or Q-M242 is a Y-chromosome DNA haplogroup. It has one primary subclade, Haplogroup Q1 (L232/S432), which includes numerous subclades that have been sampled and identified in males among modern populations.
Q-M242 is the predomina ...
(M242) (Occurred ~15,000–20,000 bp. Found in Asia and the Americas)
**** Haplogroup Q-M3
Haplogroup Q-M3 (Y-DNA) is a Y-chromosome DNA haplogroup. Haplogroup Q-M3 is a subclade of Haplogroup Q-L54. Haplogroup Q-M3 was previously known as Haplogroup Q3; currently Q-M3 is Q1b1a1a below Q1b-M346.
In 1996 the research group at Stanfo ...
(M3) (North America, Central America, and South America)
*** Haplogroup R Haplogroup R may refer to:
* Haplogroup R (mtDNA), a human mitochondrial DNA (mtDNA) haplogroup
* Haplogroup R (Y-DNA), a human Y-chromosome (Y-DNA) haplogroup
{{Disambig ...
(M207)
**** Haplogroup R1
Haplogroup R1, or R-M173, is a Y-chromosome DNA haplogroup. A primary subclade of Haplogroup R (R-M207), it is defined by the SNP M173. The other primary subclade of Haplogroup R is Haplogroup R2 (R-M479).
Males carrying R-M173 in modern populat ...
(M173)
***** Haplogroup R1a
Haplogroup R1a, or haplogroup R-M420, is a human Y-chromosome DNA haplogroup which is distributed in a large region in Eurasia, extending from Scandinavia and Central Europe to southern Siberia and South Asia.
While R1a originated c. 22,000 to ...
(M17) (Central Asia, South Asia, and Central, Northern, and Eastern Europe)
***** Haplogroup R1b (M343) (Europe, Caucasus, Central Asia, South Asia, North Africa, Central Africa)
**** Haplogroup R2
Haplogroup R2, or R-M479, is a Y-chromosome haplogroup characterized by genetic marker M479. It is one of two primary descendants of Haplogroup R (R-M207), the other being R1 (R-M173).
R-M479 has been concentrated geographically in South Asia ...
(M124) (South Asia, Caucasus, Central Asia)
** Haplogroup S (M230, P202, P204) (New Guinea, Melanesia, eastern Indonesia)
Human mitochondrial DNA haplogroups
mtDNA
Mitochondrial DNA (mtDNA or mDNA) is the DNA located in mitochondria, cellular organelles within eukaryotic cells that convert chemical energy from food into a form that cells can use, such as adenosine triphosphate (ATP). Mitochondrial DNA ...
haplogroups are lettered:
A,
B,
C,
CZ,
D,
E,
F,
G,
H,
HV,
I,
J,
pre-JT,
JT,
K,
L0,
L1,
L2,
L3,
L4,
L5,
L6,
M,
N,
P,
Q,
R,
R0,
S,
T,
U,
V,
W,
X,
Y, and
Z. The most up-to-date version of the mtDNA tree is maintained by Mannis van Oven on the PhyloTree website.
Mitochondrial Eve
In human genetics, the Mitochondrial Eve (also ''mt-Eve, mt-MRCA'') is the matrilineal most recent common ancestor (MRCA) of all living humans. In other words, she is defined as the most recent woman from whom all living humans descend in an ...
is the name given by researchers to the woman who is the most recent common matrilineal (female-lineage) ancestor of all living humans.
Defining populations
 Haplogroups can be used to define genetic populations and are often geographically oriented. For example, the following are common divisions for mtDNA haplogroups:
* African: L0, L1, L2, L3, L4, L5, L6
* West Eurasian: H, T, U, V, X, K, I, J, W (all listed West Eurasian haplogroups are derived from macro-haplogroup N)
* East Eurasian: A, B, C, D, E, F, G, Y, Z (note: C, D, E, G, and Z belong to macro-haplogroup M)
* Native American: A, B, C, D, X
*
Haplogroups can be used to define genetic populations and are often geographically oriented. For example, the following are common divisions for mtDNA haplogroups:
* African: L0, L1, L2, L3, L4, L5, L6
* West Eurasian: H, T, U, V, X, K, I, J, W (all listed West Eurasian haplogroups are derived from macro-haplogroup N)
* East Eurasian: A, B, C, D, E, F, G, Y, Z (note: C, D, E, G, and Z belong to macro-haplogroup M)
* Native American: A, B, C, D, X
* Australo-Melanesian
Australo-Melanesians (also known as Australasians or the Australomelanesoid, Australoid or Australioid race) is an outdated historical grouping of various people indigenous to Melanesia and Australia. Controversially, groups from Southeast Asia an ...
: P, Q, S
The mitochondrial haplogroups are divided into three main groups, which are designated by the sequential letters L, M, N.
Humanity first split within the L group between L0 and L1-6. L1-6 gave rise to other L groups, one of which, L3, split into the M and N group.
The M group comprises the first wave of human migration which is thought to have evolved outside of Africa, following an eastward route along southern coastal areas. Descendant lineages of haplogroup M are now found throughout Asia, the Americas, and Melanesia, as well as in parts of the Horn of Africa and North Africa; almost none have been found in Europe. The N haplogroup may represent another macrolineage that evolved outside of Africa, heading northward instead of eastward. Shortly after the migration, the large R group split off from the N.
Haplogroup R consists of two subgroups defined on the basis of their geographical distributions, one found in southeastern Asia and Oceania and the other containing almost all of the modern European populations. Haplogroup N(xR), i.e. mtDNA that belongs to the N group but not to its R subgroup, is typical of Australian aboriginal populations, while also being present at low frequencies among many populations of Eurasia and the Americas.
The L type consists of nearly all Africans.
The M type consists of:
M1 – Ethiopian, Somali and Indian populations. Likely due to much gene flow between the Horn of Africa and the Arabian Peninsula (Saudi Arabia, Yemen, Oman), separated only by a narrow strait between the Red Sea and the Gulf of Aden.
CZ – Many Siberians; branch C – Some Amerindian; branch Z – Many Saami, some Korean, some North Chinese, some Central Asian populations.
D – Some Amerindians, many Siberians and northern East Asians
E – Malay, Borneo, Philippines, Taiwanese aborigines
Taiwanese may refer to:
* Taiwanese language, another name for Taiwanese Hokkien
* Something from or related to Taiwan
Taiwan, officially the Republic of China (ROC), is a country in East Asia, at the junction of the East and South China ...
, Papua New Guinea
G – Many Northeast Siberians, northern East Asians, and Central Asians
Q – Melanesian, Polynesian, New Guinean populations
The N type consists of:
A – Found in many Amerindians and some East Asians and Siberians
I – 10% frequency in Northern, Eastern Europe
S – Some Australian aborigines
W – Some Eastern Europeans, South Asians, and southern East Asians
X – Some Amerindians, Southern Siberians, Southwest Asians, and Southern Europeans
Y – Most Nivkhs
The Nivkh, or Gilyak (also Nivkhs or Nivkhi, or Gilyaks; ethnonym: Нивхгу, ''Nʼivxgu'' (Amur) or Ниғвңгун, ''Nʼiɣvŋgun'' (E. Sakhalin) "the people"), are an indigenous ethnic group inhabiting the northern half of Sakhalin Islan ...
and people of Nias
Nias ( id, Pulau Nias, Nias language: ''Tanö Niha'') (sometimes called Little Sumatra in English) is an island located off the western coast of Sumatra, Indonesia. Nias is also the name of the archipelago () of which the island is the centre, ...
; many Ainus, Tungusic people, and Austronesians
The Austronesian peoples, sometimes referred to as Austronesian-speaking peoples, are a large group of peoples in Taiwan, Maritime Southeast Asia, Micronesia, coastal New Guinea, Island Melanesia, Polynesia, and Madagascar that speak Austrone ...
; also found with low frequency in some other populations of Siberia, East Asia, and Central Asia
R – Large group found within the N type. Populations contained therein can be divided geographically into West Eurasia and East Eurasia. Almost all European populations and a large number of Middle-Eastern population today are contained within this branch. A smaller percentage is contained in other N type groups (See above). Below are subclades of R:
B – Some Chinese, Tibetans, Mongolians, Central Asians, Koreans, Amerindians, South Siberians, Japanese, Austronesians
F – Mainly found in southeastern Asia, especially Vietnam
Vietnam or Viet Nam ( vi, Việt Nam, ), officially the Socialist Republic of Vietnam,., group="n" is a country in Southeast Asia, at the eastern edge of mainland Southeast Asia, with an area of and population of 96 million, making i ...
; 8.3% in Hvar Island in Croatia.
R0 – Found in Arabia and among Ethiopians and Somalis; branch HV (branch H; branch V) – Europe, Western Asia, North Africa;
Pre-JT – Arose in the Levant (modern Lebanon area), found in 25% frequency in Bedouin populations; branch JT (branch J; branch T) – North, Eastern Europe, Indus, Mediterranean
U – High frequency in West Eurasia, Indian sub-continent, and Algeria, found from India to the Mediterranean and to the rest of Europe; U5 in particular shows high frequency in Scandinavia and Baltic countries with the highest frequency in the Sami people.
Y-chromosome and MtDNA geographic haplogroup assignation
Here is a list of Y-chromosome and MtDNA geographic haplogroup assignation proposed by Bekada et al. 2013.Y-chromosome
According to SNPS haplogroups which are the age of the first extinction event tend to be around 45–50 kya. Haplogroups of the second extinction event seemed to diverge 32–35 kya according to Mal'ta. The ground zero extinction event appears to be Toba during which haplogroup CDEF* appeared to diverge into C, DE and F. C and F have almost nothing in common while D and E have plenty in common. Extinction event #1 according to current estimates occurred after Toba, although older ancient DNA could push the ground zero extinction event to long before Toba, and push the first extinction event here back to Toba. Haplogroups with extinction event notes by them have a dubious origin and this is because extinction events lead to severe bottlenecks, so all notes by these groups are just guesses. Note that the SNP counting of ancient DNA can be highly variable meaning that even though all these groups diverged around the same time no one knows when.mtDNA
See also
*International HapMap Project
The International HapMap Project was an organization that aimed to develop a haplotype map (HapMap) of the human genome, to describe the common patterns of human genetic variation. HapMap is used to find genetic variants affecting health, disease ...
* Molecular evolution
Molecular evolution is the process of change in the sequence composition of cellular molecules such as DNA, RNA, and proteins across generations. The field of molecular evolution uses principles of evolutionary biology and population genetics ...
* Molecular phylogenetics
Molecular phylogenetics () is the branch of phylogeny that analyzes genetic, hereditary molecular differences, predominantly in DNA sequences, to gain information on an organism's evolutionary relationships. From these analyses, it is possible to ...
* Human evolutionary genetics
Human evolutionary genetics studies how one human genome differs from another human genome, the evolutionary past that gave rise to the human genome, and its current effects. Differences between genomes have anthropological, medical, historical and ...
* Race (biology)
In biological taxonomy, race is an informal rank in the taxonomic hierarchy for which various definitions exist. Sometimes it is used to denote a level below that of subspecies, while at other times it is used as a synonym for subspecies. It has ...
/ Race (human categorization)
A race is a categorization of humans based on shared physical or social qualities into groups generally viewed as distinct within a given society. The term came into common usage during the 1500s, when it was used to refer to groups of variou ...
* Genetic genealogy
* Genealogical DNA test
A genealogical DNA test is a DNA-based test used in genetic genealogy that looks at specific locations of a person's genome in order to find or verify ancestral genealogical relationships, or (with lower reliability) to estimate the ethnic mixt ...
* List of genetic genealogy topics
This is a list of genetic genealogy topics.
Important concepts
* Genetic genealogy
* Genealogical DNA test
* Human mitochondrial DNA haplogroups
* Human Y-chromosome DNA haplogroups
* Allele
* Allele frequency
* Electropherogram
* Genetic reco ...
* List of haplogroups of notable people
References
External links
General
The Genographic Project
all DNA haplogroups
Y-Chromosome *https://web.archive.org/web/20040728005528/http://www.scs.uiuc.edu/~mcdonald/WorldHaplogroupsMaps.pdfY chromosome DNA haplogroups
Y Chromosome Consortium
PhyloTree's Y-tree
A minimal reference phylogeny for the human Y-chromosome
* The Y Chromosome Consortium (2002)
A Nomenclature System for the Tree of Human Y-Chromosomal Binary Haplogroups
Genome Research, Vol. 12(2), 339–48, February 2002. (Detailed hierarchica
chart
has conversions from previous naming schemes) * Semino et al. (2000)
The Genetic Legacy of Paleolithic Homo sapiens sapiens in Extant Europeans
Science, Vol 290 (paper which introduced the "Eu" haplogroups).
Y-DNA Ethnographic and Genographic Atlas and Open-Source Data Compilation
Mitochondrial DNA haplogroups
* PhyloTree – The phylogenetic tree of global human mitochondrial DNA variationPhyloD3 – D3.js-based phylogenetic tree based on PhyloTree
MitoTool – a web server for the analysis and retrieval of human mitochondrial DNA sequence variations
HaploGrep – automatic classification of mitochondrial DNA haplogroups based on PhyloTree
{{Webarchive, url=https://web.archive.org/web/20160612075145/http://haplogrep.uibk.ac.at/ , date=2016-06-12
HaploFind – fast automatic haplogroup assignment pipeline for human mitochondrial DNA
graphical mtDNA haplogroup skeleton
The Making of the African mtDNA Landscape
Do the Four Clades of the mtDNA Haplogroup L2 Evolve at Different Rates?
Software
Y-DNA Haplogroup Browser
DNA Human evolution Phylogenetics Population genetics Classical genetics