Deoxyribonucleic acid (; DNA) is a
polymer
A polymer () is a chemical substance, substance or material that consists of very large molecules, or macromolecules, that are constituted by many repeat unit, repeating subunits derived from one or more species of monomers. Due to their br ...
composed of two
polynucleotide
In molecular biology, a polynucleotide () is a biopolymer composed of nucleotide monomers that are covalently bonded in a chain. DNA (deoxyribonucleic acid) and RNA (ribonucleic acid) are examples of polynucleotides with distinct biological func ...
chains that coil around each other to form a
double helix
In molecular biology, the term double helix refers to the structure formed by base pair, double-stranded molecules of nucleic acids such as DNA. The double Helix, helical structure of a nucleic acid complex arises as a consequence of its Nuclei ...
. The polymer carries
genetic instructions for the development, functioning, growth and
reproduction
Reproduction (or procreation or breeding) is the biological process by which new individual organisms – "offspring" – are produced from their "parent" or parents. There are two forms of reproduction: Asexual reproduction, asexual and Sexual ...
of all known
organism
An organism is any life, living thing that functions as an individual. Such a definition raises more problems than it solves, not least because the concept of an individual is also difficult. Many criteria, few of them widely accepted, have be ...
s and many
virus
A virus is a submicroscopic infectious agent that replicates only inside the living Cell (biology), cells of an organism. Viruses infect all life forms, from animals and plants to microorganisms, including bacteria and archaea. Viruses are ...
es. DNA and
ribonucleic acid
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins ( messenger RNA). RNA and deoxyr ...
(RNA) are
nucleic acid
Nucleic acids are large biomolecules that are crucial in all cells and viruses. They are composed of nucleotides, which are the monomer components: a pentose, 5-carbon sugar, a phosphate group and a nitrogenous base. The two main classes of nuclei ...
s. Alongside
protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
s,
lipids
Lipids are a broad group of organic compounds which include fats, waxes, sterols, fat-soluble vitamins (such as vitamins Vitamin A, A, Vitamin D, D, Vitamin E, E and Vitamin K, K), monoglycerides, diglycerides, phospholipids, and others. The fu ...
and complex carbohydrates (
polysaccharide
Polysaccharides (), or polycarbohydrates, are the most abundant carbohydrates found in food. They are long-chain polymeric carbohydrates composed of monosaccharide units bound together by glycosidic linkages. This carbohydrate can react with wat ...
s), nucleic acids are one of the four major types of
macromolecule
A macromolecule is a "molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass." Polymers are physi ...
s that are essential for all known forms of
life
Life, also known as biota, refers to matter that has biological processes, such as Cell signaling, signaling and self-sustaining processes. It is defined descriptively by the capacity for homeostasis, Structure#Biological, organisation, met ...
.
The two DNA strands are known as polynucleotides as they are composed of simpler
monomer
A monomer ( ; ''mono-'', "one" + '' -mer'', "part") is a molecule that can react together with other monomer molecules to form a larger polymer chain or two- or three-dimensional network in a process called polymerization.
Classification
Chemis ...
ic units called
nucleotide
Nucleotides are Organic compound, organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both o ...
s. Each nucleotide is composed of one of four
nitrogen-containing nucleobase
Nucleotide bases (also nucleobases, nitrogenous bases) are nitrogen-containing biological compounds that form nucleosides, which, in turn, are components of nucleotides, with all of these monomers constituting the basic building blocks of nuc ...
s (
cytosine
Cytosine () (symbol C or Cyt) is one of the four nucleotide bases found in DNA and RNA, along with adenine, guanine, and thymine ( uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attac ...
guanine
Guanine () (symbol G or Gua) is one of the four main nucleotide bases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine ( uracil in RNA). In DNA, guanine is paired with cytosine. The guanine nucleoside ...
adenine
Adenine (, ) (nucleoside#List of nucleosides and corresponding nucleobases, symbol A or Ade) is a purine nucleotide base that is found in DNA, RNA, and Adenosine triphosphate, ATP. Usually a white crystalline subtance. The shape of adenine is ...
or
thymine
Thymine () (symbol T or Thy) is one of the four nucleotide bases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The others are adenine, guanine, and cytosine. Thymine is also known as 5-methyluracil, a pyrimidine ...
, a
sugar
Sugar is the generic name for sweet-tasting, soluble carbohydrates, many of which are used in food. Simple sugars, also called monosaccharides, include glucose
Glucose is a sugar with the Chemical formula#Molecular formula, molecul ...
called
deoxyribose
Deoxyribose, or more precisely 2-deoxyribose, is a monosaccharide with idealized formula H−(C=O)−(CH2)−(CHOH)3−H. Its name indicates that it is a deoxy sugar, meaning that it is derived from the sugar ribose by loss of a hydroxy group. D ...
, and a
phosphate group
Phosphates are the naturally occurring form of the element phosphorus.
In chemistry, a phosphate is an anion, salt, functional group or ester derived from a phosphoric acid. It most commonly means orthophosphate, a derivative of orthophosp ...
. The nucleotides are joined to one another in a chain by
covalent bond
A covalent bond is a chemical bond that involves the sharing of electrons to form electron pairs between atoms. These electron pairs are known as shared pairs or bonding pairs. The stable balance of attractive and repulsive forces between atom ...
s (known as the
phosphodiester linkage
In chemistry, a phosphodiester bond occurs when exactly two of the hydroxyl groups () in phosphoric acid react with hydroxyl groups on other molecules to form two ester bonds. The "bond" involves this linkage . Discussion of phosphodiesters is d ...
) between the sugar of one nucleotide and the phosphate of the next, resulting in an alternating
sugar-phosphate backbone. The nitrogenous bases of the two separate polynucleotide strands are bound together, according to
base pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
ing rules (A with T and C with G), with
hydrogen bond
In chemistry, a hydrogen bond (H-bond) is a specific type of molecular interaction that exhibits partial covalent character and cannot be described as a purely electrostatic force. It occurs when a hydrogen (H) atom, Covalent bond, covalently b ...
s to make double-stranded DNA. The complementary nitrogenous bases are divided into two groups, the single-ringed
pyrimidine
Pyrimidine (; ) is an aromatic, heterocyclic, organic compound similar to pyridine (). One of the three diazines (six-membered heterocyclics with two nitrogen atoms in the ring), it has nitrogen atoms at positions 1 and 3 in the ring. The oth ...
s and the double-ringed
purine
Purine is a heterocyclic aromatic organic compound that consists of two rings (pyrimidine and imidazole) fused together. It is water-soluble. Purine also gives its name to the wider class of molecules, purines, which include substituted puri ...
s. In DNA, the pyrimidines are thymine and cytosine; the purines are adenine and guanine.
Both strands of double-stranded DNA store the same
biological information. This information is
replicated when the two strands separate. A large part of DNA (more than 98% for humans) is
non-coding
Non-coding DNA (ncDNA) sequences are components of an organism's DNA that do not encode protein sequences. Some non-coding DNA is transcribed into functional non-coding RNA molecules (e.g. transfer RNA, microRNA, piRNA, ribosomal RNA, and regula ...
, meaning that these sections do not serve as patterns for
protein sequences
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthes ...
. The two strands of DNA run in opposite directions to each other and are thus
antiparallel. Attached to each sugar is one of four types of nucleobases (or ''bases''). It is the
sequence
In mathematics, a sequence is an enumerated collection of objects in which repetitions are allowed and order matters. Like a set, it contains members (also called ''elements'', or ''terms''). The number of elements (possibly infinite) is cal ...
of these four nucleobases along the backbone that encodes genetic information. RNA strands are created using DNA strands as a template in a process called
transcription, where DNA bases are exchanged for their corresponding bases except in the case of thymine (T), for which RNA substitutes
uracil
Uracil () (nucleoside#List of nucleosides and corresponding nucleobases, symbol U or Ura) is one of the four nucleotide bases in the nucleic acid RNA. The others are adenine (A), cytosine (C), and guanine (G). In RNA, uracil binds to adenine via ...
(U). Under the
genetic code
Genetic code is a set of rules used by living cell (biology), cells to Translation (biology), translate information encoded within genetic material (DNA or RNA sequences of nucleotide triplets or codons) into proteins. Translation is accomplished ...
, these RNA strands specify the sequence of
amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
s within proteins in a process called
translation
Translation is the communication of the semantics, meaning of a #Source and target languages, source-language text by means of an Dynamic and formal equivalence, equivalent #Source and target languages, target-language text. The English la ...
.
Within eukaryotic cells, DNA is organized into long structures called
chromosome
A chromosome is a package of DNA containing part or all of the genetic material of an organism. In most chromosomes, the very long thin DNA fibers are coated with nucleosome-forming packaging proteins; in eukaryotic cells, the most import ...
s. Before typical
cell division
Cell division is the process by which a parent cell (biology), cell divides into two daughter cells. Cell division usually occurs as part of a larger cell cycle in which the cell grows and replicates its chromosome(s) before dividing. In eukar ...
, these chromosomes are duplicated in the process of DNA replication, providing a complete set of chromosomes for each daughter cell.
Eukaryotic organisms (
animal
Animals are multicellular, eukaryotic organisms in the Biology, biological Kingdom (biology), kingdom Animalia (). With few exceptions, animals heterotroph, consume organic material, Cellular respiration#Aerobic respiration, breathe oxygen, ...
s,
plant
Plants are the eukaryotes that form the Kingdom (biology), kingdom Plantae; they are predominantly Photosynthesis, photosynthetic. This means that they obtain their energy from sunlight, using chloroplasts derived from endosymbiosis with c ...
s,
fungi
A fungus (: fungi , , , or ; or funguses) is any member of the group of eukaryotic organisms that includes microorganisms such as yeasts and mold (fungus), molds, as well as the more familiar mushrooms. These organisms are classified as one ...
and
protist
A protist ( ) or protoctist is any eukaryotic organism that is not an animal, land plant, or fungus. Protists do not form a natural group, or clade, but are a paraphyletic grouping of all descendants of the last eukaryotic common ancest ...
s) store most of their DNA inside the
cell nucleus
The cell nucleus (; : nuclei) is a membrane-bound organelle found in eukaryote, eukaryotic cell (biology), cells. Eukaryotic cells usually have a single nucleus, but a few cell types, such as mammalian red blood cells, have #Anucleated_cells, ...
as
nuclear DNA
Nuclear DNA (nDNA), or nuclear deoxyribonucleic acid, is the DNA contained within each cell nucleus of a eukaryotic organism. It encodes for the majority of the genome in eukaryotes, with mitochondrial DNA and plastid DNA coding for the rest. ...
, and some in the
mitochondria
A mitochondrion () is an organelle found in the cells of most eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is us ...
as
mitochondrial DNA
Mitochondrial DNA (mtDNA and mDNA) is the DNA located in the mitochondrion, mitochondria organelles in a eukaryotic cell that converts chemical energy from food into adenosine triphosphate (ATP). Mitochondrial DNA is a small portion of the D ...
or in
chloroplast
A chloroplast () is a type of membrane-bound organelle, organelle known as a plastid that conducts photosynthesis mostly in plant cell, plant and algae, algal cells. Chloroplasts have a high concentration of chlorophyll pigments which captur ...
s as
chloroplast DNA
Chloroplast DNA (cpDNA), also known as plastid DNA (ptDNA) is the DNA located in chloroplasts, which are photosynthetic organelles located within the cells of some eukaryotic organisms. Chloroplasts, like other types of plastid, contain a genome s ...
. In contrast,
prokaryote
A prokaryote (; less commonly spelled procaryote) is a unicellular organism, single-celled organism whose cell (biology), cell lacks a cell nucleus, nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Ancient Gree ...
s (
bacteria
Bacteria (; : bacterium) are ubiquitous, mostly free-living organisms often consisting of one Cell (biology), biological cell. They constitute a large domain (biology), domain of Prokaryote, prokaryotic microorganisms. Typically a few micr ...
and
archaea
Archaea ( ) is a Domain (biology), domain of organisms. Traditionally, Archaea only included its Prokaryote, prokaryotic members, but this has since been found to be paraphyletic, as eukaryotes are known to have evolved from archaea. Even thou ...
) store their DNA only in the
cytoplasm
The cytoplasm describes all the material within a eukaryotic or prokaryotic cell, enclosed by the cell membrane, including the organelles and excluding the nucleus in eukaryotic cells. The material inside the nucleus of a eukaryotic cell a ...
, in
circular chromosome
A circular chromosome is a chromosome in bacteria, archaea, mitochondria, and chloroplasts, in the form of a molecule of circular DNA, unlike the linear chromosome of most eukaryotes.
Most prokaryote chromosomes contain a circular DNA molecule. ...
s. Within eukaryotic chromosomes,
chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important r ...
proteins, such as
histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei and in most Archaeal phyla. They act as spools around which DNA winds to create structural units called nucleosomes ...
s, compact and organize DNA. These compacting structures guide the interactions between DNA and other proteins, helping control which parts of the DNA are transcribed.
Properties

DNA is a long
polymer
A polymer () is a chemical substance, substance or material that consists of very large molecules, or macromolecules, that are constituted by many repeat unit, repeating subunits derived from one or more species of monomers. Due to their br ...
made from repeating units called
nucleotide
Nucleotides are Organic compound, organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both o ...
s.
The structure of DNA is dynamic along its length, being capable of coiling into tight loops and other shapes. In all species it is composed of two helical chains, bound to each other by
hydrogen bonds
In chemistry, a hydrogen bond (H-bond) is a specific type of molecular interaction that exhibits partial covalent character and cannot be described as a purely electrostatic force. It occurs when a hydrogen (H) atom, covalently bonded to a mo ...
. Both chains are coiled around the same axis, and have the same
pitch of . The pair of chains have a radius of .
According to another study, when measured in a different solution, the DNA chain measured wide, and one nucleotide unit measured long. The buoyant density of most DNA is 1.7g/cm
3.
DNA does not usually exist as a single strand, but instead as a pair of strands that are held tightly together.
These two long strands coil around each other, in the shape of a
double helix
In molecular biology, the term double helix refers to the structure formed by base pair, double-stranded molecules of nucleic acids such as DNA. The double Helix, helical structure of a nucleic acid complex arises as a consequence of its Nuclei ...
. The nucleotide contains both a segment of the
backbone
The spinal column, also known as the vertebral column, spine or backbone, is the core part of the axial skeleton in vertebrates. The vertebral column is the defining and eponymous characteristic of the vertebrate. The spinal column is a segmente ...
of the molecule (which holds the chain together) and a
nucleobase
Nucleotide bases (also nucleobases, nitrogenous bases) are nitrogen-containing biological compounds that form nucleosides, which, in turn, are components of nucleotides, with all of these monomers constituting the basic building blocks of nuc ...
(which interacts with the other DNA strand in the helix). A nucleobase linked to a sugar is called a
nucleoside
Nucleosides are glycosylamines that can be thought of as nucleotides without a phosphate group. A nucleoside consists simply of a nucleobase (also termed a nitrogenous base) and a five-carbon sugar (ribose or 2'-deoxyribose) whereas a nucleotid ...
, and a base linked to a sugar and to one or more phosphate groups is called a
nucleotide
Nucleotides are Organic compound, organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both o ...
. A
biopolymer
Biopolymers are natural polymers produced by the cells of living organisms. Like other polymers, biopolymers consist of monomeric units that are covalently bonded in chains to form larger molecules. There are three main classes of biopolymers, ...
comprising multiple linked nucleotides (as in DNA) is called a
polynucleotide
In molecular biology, a polynucleotide () is a biopolymer composed of nucleotide monomers that are covalently bonded in a chain. DNA (deoxyribonucleic acid) and RNA (ribonucleic acid) are examples of polynucleotides with distinct biological func ...
.
The backbone of the DNA strand is made from alternating
phosphate
Phosphates are the naturally occurring form of the element phosphorus.
In chemistry, a phosphate is an anion, salt, functional group or ester derived from a phosphoric acid. It most commonly means orthophosphate, a derivative of orthop ...
and
sugar
Sugar is the generic name for sweet-tasting, soluble carbohydrates, many of which are used in food. Simple sugars, also called monosaccharides, include glucose
Glucose is a sugar with the Chemical formula#Molecular formula, molecul ...
groups.
The sugar in DNA is
2-deoxyribose, which is a
pentose
In chemistry, a pentose is a monosaccharide (simple sugar) with five carbon atoms. The chemical formula of many pentoses is , and their molecular weight is 150.13 g/mol.[carbon
Carbon () is a chemical element; it has chemical symbol, symbol C and atomic number 6. It is nonmetallic and tetravalence, tetravalent—meaning that its atoms are able to form up to four covalent bonds due to its valence shell exhibiting 4 ...]
) sugar. The sugars are joined by phosphate groups that form
phosphodiester bond
In chemistry, a phosphodiester bond occurs when exactly two of the hydroxyl groups () in phosphoric acid react with hydroxyl groups on other molecules to form two ester bonds. The "bond" involves this linkage . Discussion of phosphodiesters is d ...
s between the third and fifth carbon
atom
Atoms are the basic particles of the chemical elements. An atom consists of a atomic nucleus, nucleus of protons and generally neutrons, surrounded by an electromagnetically bound swarm of electrons. The chemical elements are distinguished fr ...
s of adjacent sugar rings. These are known as the
3′-end (three prime end), and
5′-end (five prime end) carbons, the prime symbol being used to distinguish these carbon atoms from those of the base to which the deoxyribose forms a
glycosidic bond
A glycosidic bond or glycosidic linkage is a type of ether bond that joins a carbohydrate (sugar) molecule to another group, which may or may not be another carbohydrate.
A glycosidic bond is formed between the hemiacetal or hemiketal group o ...
.
Therefore, any DNA strand normally has one end at which there is a phosphate group attached to the 5′ carbon of a ribose (the 5′ phosphoryl) and another end at which there is a free hydroxyl group attached to the 3′ carbon of a ribose (the 3′ hydroxyl). The orientation of the 3′ and 5′ carbons along the sugar-phosphate backbone confers
directionality (sometimes called polarity) to each DNA strand. In a
nucleic acid double helix
In molecular biology, the term double helix refers to the structure formed by double-stranded molecules of nucleic acids such as DNA. The double helical structure of a nucleic acid complex arises as a consequence of its secondary structure, a ...
, the direction of the nucleotides in one strand is opposite to their direction in the other strand: the strands are
antiparallel. The asymmetric ends of DNA strands are said to have a directionality of five prime end (5′ ), and three prime end (3′), with the 5′ end having a terminal phosphate group and the 3′ end a terminal hydroxyl group. One major difference between DNA and
RNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA and deoxyrib ...
is the sugar, with the 2-deoxyribose in DNA being replaced by the related pentose sugar
ribose
Ribose is a simple sugar and carbohydrate with molecular formula C5H10O5 and the linear-form composition H−(C=O)−(CHOH)4−H. The naturally occurring form, , is a component of the ribonucleotides from which RNA is built, and so this comp ...
in RNA.

The DNA double helix is stabilized primarily by two forces:
hydrogen bond
In chemistry, a hydrogen bond (H-bond) is a specific type of molecular interaction that exhibits partial covalent character and cannot be described as a purely electrostatic force. It occurs when a hydrogen (H) atom, Covalent bond, covalently b ...
s between nucleotides and
base-stacking interactions among
aromatic
In organic chemistry, aromaticity is a chemical property describing the way in which a conjugated system, conjugated ring of unsaturated bonds, lone pairs, or empty orbitals exhibits a stabilization stronger than would be expected from conjugati ...
nucleobases.
The four bases found in DNA are
adenine
Adenine (, ) (nucleoside#List of nucleosides and corresponding nucleobases, symbol A or Ade) is a purine nucleotide base that is found in DNA, RNA, and Adenosine triphosphate, ATP. Usually a white crystalline subtance. The shape of adenine is ...
(),
cytosine
Cytosine () (symbol C or Cyt) is one of the four nucleotide bases found in DNA and RNA, along with adenine, guanine, and thymine ( uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attac ...
(),
guanine
Guanine () (symbol G or Gua) is one of the four main nucleotide bases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine ( uracil in RNA). In DNA, guanine is paired with cytosine. The guanine nucleoside ...
() and
thymine
Thymine () (symbol T or Thy) is one of the four nucleotide bases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The others are adenine, guanine, and cytosine. Thymine is also known as 5-methyluracil, a pyrimidine ...
(). These four bases are attached to the sugar-phosphate to form the complete nucleotide, as shown for
adenosine monophosphate
Adenosine monophosphate (AMP), also known as 5'-adenylic acid, is a nucleotide. AMP consists of a phosphate group, the sugar ribose, and the nucleobase adenine. It is an ester of phosphoric acid and the nucleoside adenosine. As a substituent it t ...
. Adenine pairs with thymine and guanine pairs with cytosine, forming and
base pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
s.
Nucleobase classification
The nucleobases are classified into two types: the
purine
Purine is a heterocyclic aromatic organic compound that consists of two rings (pyrimidine and imidazole) fused together. It is water-soluble. Purine also gives its name to the wider class of molecules, purines, which include substituted puri ...
s, and , which are fused five- and six-membered
heterocyclic compound
A heterocyclic compound or ring structure is a cyclic compound that has atoms of at least two different elements as members of its ring(s). Heterocyclic organic chemistry is the branch of organic chemistry dealing with the synthesis, proper ...
s, and the
pyrimidine
Pyrimidine (; ) is an aromatic, heterocyclic, organic compound similar to pyridine (). One of the three diazines (six-membered heterocyclics with two nitrogen atoms in the ring), it has nitrogen atoms at positions 1 and 3 in the ring. The oth ...
s, the six-membered rings and .
A fifth pyrimidine nucleobase,
uracil
Uracil () (nucleoside#List of nucleosides and corresponding nucleobases, symbol U or Ura) is one of the four nucleotide bases in the nucleic acid RNA. The others are adenine (A), cytosine (C), and guanine (G). In RNA, uracil binds to adenine via ...
(), usually takes the place of thymine in RNA and differs from thymine by lacking a
methyl group
In organic chemistry, a methyl group is an alkyl derived from methane, containing one carbon atom bonded to three hydrogen atoms, having chemical formula (whereas normal methane has the formula ). In formulas, the group is often abbreviated a ...
on its ring. In addition to RNA and DNA, many artificial
nucleic acid analogue
Nucleic acid analogues are compounds which are analogous (structurally similar) to naturally occurring RNA and DNA, used in medicine and in molecular biology research. Nucleic acids are chains of nucleotides, which are composed of three parts: ...
s have been created to study the properties of nucleic acids, or for use in biotechnology.
Non-canonical bases
Modified bases occur in DNA. The first of these recognized was
5-methylcytosine
5-Methylcytosine (5mC) is a methylation, methylated form of the DNA base cytosine (C) that regulates gene Transcription (genetics), transcription and takes several other biological roles. When cytosine is methylated, the DNA maintains the same s ...
, which was found in the
genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as ...
of ''
Mycobacterium tuberculosis
''Mycobacterium tuberculosis'' (M. tb), also known as Koch's bacillus, is a species of pathogenic bacteria in the family Mycobacteriaceae and the causative agent of tuberculosis.
First discovered in 1882 by Robert Koch, ''M. tuberculosis'' ha ...
'' in 1925.
The reason for the presence of these noncanonical bases in bacterial viruses (
bacteriophage
A bacteriophage (), also known informally as a phage (), is a virus that infects and replicates within bacteria. The term is derived . Bacteriophages are composed of proteins that Capsid, encapsulate a DNA or RNA genome, and may have structu ...
s) is to avoid the
restriction enzyme
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class o ...
s present in bacteria. This enzyme system acts at least in part as a molecular immune system protecting bacteria from infection by viruses.
Modifications of the bases cytosine and adenine, the more common and modified DNA bases, play vital roles in the
epigenetic
In biology, epigenetics is the study of changes in gene expression that happen without changes to the DNA sequence. The Greek prefix ''epi-'' (ἐπι- "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in ...
control of gene expression in plants and animals.
A number of noncanonical bases are known to occur in DNA.
Most of these are modifications of the canonical bases plus uracil.
* Modified Adenine
** N6-carbamoyl-methyladenine
** N6-methyadenine
* Modified Guanine
** 7-Deazaguanine
** 7-Methylguanine
* Modified Cytosine
** N4-Methylcytosine
** 5-Carboxylcytosine
** 5-Formylcytosine
** 5-Glycosylhydroxymethylcytosine
** 5-Hydroxycytosine
** 5-Methylcytosine
* Modified Thymidine
** α-Glutamythymidine
** α-Putrescinylthymine
* Uracil and modifications
**
Base J
β-D-Glucopyranosyloxymethyluracil or base J is a hypermodified nucleobase found in the DNA of kinetoplastids including the human Pathogen, pathogenic trypanosomes. It was discovered in 1993, in the trypanosome ''Trypanosoma brucei'' and was the f ...
** Uracil
** 5-Dihydroxypentauracil
** 5-Hydroxymethyldeoxyuracil
* Others
** Deoxyarchaeosine
** 2,6-Diaminopurine (2-Aminoadenine)
Grooves

Twin helical strands form the DNA backbone. Another double helix may be found tracing the spaces, or grooves, between the strands. These voids are adjacent to the base pairs and may provide a
binding site
In biochemistry and molecular biology, a binding site is a region on a macromolecule such as a protein that binds to another molecule with specificity. The binding partner of the macromolecule is often referred to as a ligand. Ligands may includ ...
. As the strands are not symmetrically located with respect to each other, the grooves are unequally sized. The major groove is wide, while the minor groove is in width. Due to the larger width of the major groove, the edges of the bases are more accessible in the major groove than in the minor groove. As a result, proteins such as
transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription (genetics), transcription of genetics, genetic information from DNA to messenger RNA, by binding t ...
s that can bind to specific sequences in double-stranded DNA usually make contact with the sides of the bases exposed in the major groove.
This situation varies in unusual conformations of DNA within the cell ''(see below)'', but the major and minor grooves are always named to reflect the differences in width that would be seen if the DNA was twisted back into the ordinary
B form.
Base pairing
Top, a base pair with three
hydrogen bond
In chemistry, a hydrogen bond (H-bond) is a specific type of molecular interaction that exhibits partial covalent character and cannot be described as a purely electrostatic force. It occurs when a hydrogen (H) atom, Covalent bond, covalently b ...
s. Bottom, an base pair with two hydrogen bonds. Non-covalent hydrogen bonds between the pairs are shown as dashed lines.
In a DNA double helix, each type of nucleobase on one strand bonds with just one type of nucleobase on the other strand. This is called
complementary
Complement may refer to:
The arts
* Complement (music), an interval that, when added to another, spans an octave
** Aggregate complementation, the separation of pitch-class collections into complementary sets
* Complementary color, in the visu ...
base pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
ing. Purines form
hydrogen bond
In chemistry, a hydrogen bond (H-bond) is a specific type of molecular interaction that exhibits partial covalent character and cannot be described as a purely electrostatic force. It occurs when a hydrogen (H) atom, Covalent bond, covalently b ...
s to pyrimidines, with adenine bonding only to thymine in two hydrogen bonds, and cytosine bonding only to guanine in three hydrogen bonds. This arrangement of two nucleotides binding together across the double helix (from six-carbon ring to six-carbon ring) is called a Watson-Crick base pair. DNA with high
GC-content
In molecular biology and genetics, GC-content (or guanine-cytosine content) is the percentage of nitrogenous bases in a DNA or RNA molecule that are either guanine (G) or cytosine (C). This measure indicates the proportion of G and C bases out of ...
is more stable than DNA with low -content. A
Hoogsteen base pair
A Hoogsteen base pair is a variation of base-pairing in nucleic acids such as the A•T pair. In this manner, two nucleobases, one on each strand, can be held together by hydrogen bonds in the major groove. A Hoogsteen base pair applies the N7 po ...
(hydrogen bonding the 6-carbon ring to the 5-carbon ring) is a rare variation of base-pairing.
As hydrogen bonds are not
covalent
A covalent bond is a chemical bond that involves the sharing of electrons to form electron pairs between atoms. These electron pairs are known as shared pairs or bonding pairs. The stable balance of attractive and repulsive forces between atom ...
, they can be broken and rejoined relatively easily. The two strands of DNA in a double helix can thus be pulled apart like a zipper, either by a mechanical force or high
temperature
Temperature is a physical quantity that quantitatively expresses the attribute of hotness or coldness. Temperature is measurement, measured with a thermometer. It reflects the average kinetic energy of the vibrating and colliding atoms making ...
. As a result of this base pair complementarity, all the information in the double-stranded sequence of a DNA helix is duplicated on each strand, which is vital in DNA replication. This reversible and specific interaction between complementary base pairs is critical for all the functions of DNA in organisms.
ssDNA vs. dsDNA
Most DNA molecules are actually two polymer strands, bound together in a helical fashion by noncovalent bonds; this double-stranded (dsDNA) structure is maintained largely by the intrastrand base stacking interactions, which are strongest for stacks. The two strands can come apart—a process known as melting—to form two single-stranded DNA (ssDNA) molecules. Melting occurs at high temperatures, low salt and high
pH (low pH also melts DNA, but since DNA is unstable due to acid depurination, low pH is rarely used).
The stability of the dsDNA form depends not only on the -content (% basepairs) but also on sequence (since stacking is sequence specific) and also length (longer molecules are more stable). The stability can be measured in various ways; a common way is the
melting temperature (also called ''T
m'' value), which is the temperature at which 50% of the double-strand molecules are converted to single-strand molecules; melting temperature is dependent on ionic strength and the concentration of DNA. As a result, it is both the percentage of base pairs and the overall length of a DNA double helix that determines the strength of the association between the two strands of DNA. Long DNA helices with a high -content have more strongly interacting strands, while short helices with high content have more weakly interacting strands. In biology, parts of the DNA double helix that need to separate easily, such as the
Pribnow box
The Pribnow box (also known as the Pribnow-Schaller box) is a sequence of ''TATAAT'' of six nucleotides (thymine, adenine, thymine, etc.) that is an essential part of a promoter site on DNA for transcription to occur in bacteria.
It is an ideal ...
in some
promoters, tend to have a high content, making the strands easier to pull apart.
In the laboratory, the strength of this interaction can be measured by finding the melting temperature ''T
m'' necessary to break half of the hydrogen bonds. When all the base pairs in a DNA double helix melt, the strands separate and exist in solution as two entirely independent molecules. These single-stranded DNA molecules have no single common shape, but some conformations are more stable than others.
Amount

In humans, the total female
diploid
Ploidy () is the number of complete sets of chromosomes in a cell, and hence the number of possible alleles for autosomal and pseudoautosomal genes. Here ''sets of chromosomes'' refers to the number of maternal and paternal chromosome copies, ...
nuclear genome
Nuclear DNA (nDNA), or nuclear deoxyribonucleic acid, is the DNA contained within each cell nucleus of a eukaryotic organism. It encodes for the majority of the genome in eukaryotes, with mitochondrial DNA and plastid DNA coding for the rest. It ...
per cell extends for 6.37 Gigabase pairs (Gbp), is 208.23 cm long and weighs 6.51 picograms (pg).
Male values are 6.27 Gbp, 205.00 cm, 6.41 pg.
Each DNA polymer can contain hundreds of millions of nucleotides, such as in
chromosome 1
Chromosome 1 is the designation for the largest human chromosome. Humans have two copies of chromosome 1, as they do with all of the autosomes, which are the non-sex chromosomes. Chromosome 1 spans about 249 million nucleotide base pairs, which a ...
. Chromosome 1 is the largest human
chromosome
A chromosome is a package of DNA containing part or all of the genetic material of an organism. In most chromosomes, the very long thin DNA fibers are coated with nucleosome-forming packaging proteins; in eukaryotic cells, the most import ...
with approximately 220 million
base pair
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA ...
s, and would be long if straightened.
In
eukaryote
The eukaryotes ( ) constitute the Domain (biology), domain of Eukaryota or Eukarya, organisms whose Cell (biology), cells have a membrane-bound cell nucleus, nucleus. All animals, plants, Fungus, fungi, seaweeds, and many unicellular organisms ...
s, in addition to
nuclear DNA
Nuclear DNA (nDNA), or nuclear deoxyribonucleic acid, is the DNA contained within each cell nucleus of a eukaryotic organism. It encodes for the majority of the genome in eukaryotes, with mitochondrial DNA and plastid DNA coding for the rest. ...
, there is also
mitochondrial DNA
Mitochondrial DNA (mtDNA and mDNA) is the DNA located in the mitochondrion, mitochondria organelles in a eukaryotic cell that converts chemical energy from food into adenosine triphosphate (ATP). Mitochondrial DNA is a small portion of the D ...
(mtDNA) which encodes certain proteins used by the mitochondria. The mtDNA is usually relatively small in comparison to the nuclear DNA. For example, the
human mitochondrial DNA
Human mitochondrial genetics is the study of the genetics of human mitochondrial DNA (the DNA contained in human mitochondria). The human mitochondrial genome is the entirety of hereditary information contained in human mitochondria. Mitochondr ...
forms closed circular molecules, each of which contains 16,569
DNA base pairs,
with each such molecule normally containing a full set of the mitochondrial genes. Each human mitochondrion contains, on average, approximately 5 such mtDNA molecules.
[ Each human ]cell
Cell most often refers to:
* Cell (biology), the functional basic unit of life
* Cellphone, a phone connected to a cellular network
* Clandestine cell, a penetration-resistant form of a secret or outlawed organization
* Electrochemical cell, a de ...
contains approximately 100 mitochondria, giving a total number of mtDNA molecules per human cell of approximately 500.[ However, the amount of mitochondria per cell also varies by cell type, and an ]egg cell
The egg cell or ovum (: ova) is the female Reproduction, reproductive cell, or gamete, in most anisogamous organisms (organisms that reproduce sexually with a larger, female gamete and a smaller, male one). The term is used when the female game ...
can contain 100,000 mitochondria, corresponding to up to 1,500,000 copies of the mitochondrial genome (constituting up to 90% of the DNA of the cell).
Sense and antisense
A DNA sequence
A nucleic acid sequence is a succession of bases within the nucleotides forming alleles within a DNA (using GACT) or RNA (GACU) molecule. This succession is denoted by a series of a set of five different letters that indicate the order of the nu ...
is called a "sense" sequence if it is the same as that of a messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
copy that is translated into protein. The sequence on the opposite strand is called the "antisense" sequence. Both sense and antisense sequences can exist on different parts of the same strand of DNA (i.e. both strands can contain both sense and antisense sequences). In both prokaryotes and eukaryotes, antisense RNA sequences are produced, but the functions of these RNAs are not entirely clear. One proposal is that antisense RNAs are involved in regulating gene expression
Gene expression is the process (including its Regulation of gene expression, regulation) by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, proteins or non-coding RNA, ...
through RNA-RNA base pairing.
A few DNA sequences in prokaryotes and eukaryotes, and more in plasmid
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria and ...
s and virus
A virus is a submicroscopic infectious agent that replicates only inside the living Cell (biology), cells of an organism. Viruses infect all life forms, from animals and plants to microorganisms, including bacteria and archaea. Viruses are ...
es, blur the distinction between sense and antisense strands by having overlapping gene
An overlapping gene (or OLG) is a gene whose expressible nucleotide sequence partially overlaps with the expressible nucleotide sequence of another gene. In this way, a nucleotide sequence may make a contribution to the function of one or more g ...
s. In these cases, some DNA sequences do double duty, encoding one protein when read along one strand, and a second protein when read in the opposite direction along the other strand. In bacteria
Bacteria (; : bacterium) are ubiquitous, mostly free-living organisms often consisting of one Cell (biology), biological cell. They constitute a large domain (biology), domain of Prokaryote, prokaryotic microorganisms. Typically a few micr ...
, this overlap may be involved in the regulation of gene transcription, while in viruses, overlapping genes increase the amount of information that can be encoded within the small viral genome.
Supercoiling
DNA can be twisted like a rope in a process called DNA supercoil
DNA supercoiling refers to the amount of twist in a particular DNA strand, which determines the amount of strain on it. A given strand may be "positively supercoiled" or "negatively supercoiled" (more or less tightly wound). The amount of a st ...
ing. With DNA in its "relaxed" state, a strand usually circles the axis of the double helix once every 10.4 base pairs, but if the DNA is twisted the strands become more tightly or more loosely wound. If the DNA is twisted in the direction of the helix, this is positive supercoiling, and the bases are held more tightly together. If they are twisted in the opposite direction, this is negative supercoiling, and the bases come apart more easily. In nature, most DNA has slight negative supercoiling that is introduced by enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
s called topoisomerase
DNA topoisomerases (or topoisomerases) are enzymes that catalyze changes in the topological state of DNA, interconverting relaxed and supercoiled forms, linked (catenated) and unlinked species, and knotted and unknotted DNA. Topological issues in ...
s.DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all life, living organisms, acting as the most essential part of heredity, biolog ...
.
Alternative DNA structures
 DNA exists in many possible conformations that include
DNA exists in many possible conformations that include A-DNA
A-DNA is one of the possible double helical structures which DNA can adopt. A-DNA is thought to be one of three biologically active double helical structures along with B-DNA and Z-DNA. It is a right-handed double helix fairly similar to the m ...
, B-DNA
In molecular biology, the term double helix refers to the structure formed by double-stranded molecules of nucleic acids such as DNA. The double helical structure of a nucleic acid complex arises as a consequence of its secondary structure, a ...
, and Z-DNA
Z-DNA is one of the many possible double helical structures of DNA. It is a left-handed double helical structure in which the helix winds to the left in a zigzag pattern, instead of to the right, like the more common B-DNA form. Z-DNA is thought ...
forms, although only B-DNA and Z-DNA have been directly observed in functional organisms.ion
An ion () is an atom or molecule with a net electrical charge. The charge of an electron is considered to be negative by convention and this charge is equal and opposite to the charge of a proton, which is considered to be positive by convent ...
s, and the presence of polyamine
A polyamine is an organic compound having two or more amino groups. Alkyl polyamines occur naturally, but some are synthetic. Alkylpolyamines are colorless, hygroscopic, and water soluble. Near neutral pH, they exist as the ammonium derivatives. ...
s in solution.
The first published reports of A-DNA X-ray diffraction patterns—and also B-DNA—used analyses based on Patterson function The Patterson function is used to solve the phase problem in X-ray crystallography
X-ray crystallography is the experimental science of determining the atomic and molecular structure of a crystal, in which the crystalline structure causes a beam ...
s that provided only a limited amount of structural information for oriented fibers of DNA.in vivo
Studies that are ''in vivo'' (Latin for "within the living"; often not italicized in English) are those in which the effects of various biological entities are tested on whole, living organisms or cells, usually animals, including humans, an ...
'' B-DNA X-ray diffraction-scattering patterns of highly hydrated DNA fibers in terms of squares of Bessel function
Bessel functions, named after Friedrich Bessel who was the first to systematically study them in 1824, are canonical solutions of Bessel's differential equation
x^2 \frac + x \frac + \left(x^2 - \alpha^2 \right)y = 0
for an arbitrary complex ...
s.James Watson
James Dewey Watson (born April 6, 1928) is an American molecular biology, molecular biologist, geneticist, and zoologist. In 1953, he co-authored with Francis Crick the academic paper in ''Nature (journal), Nature'' proposing the Nucleic acid ...
and Francis Crick
Francis Harry Compton Crick (8 June 1916 – 28 July 2004) was an English molecular biologist, biophysicist, and neuroscientist. He, James Watson, Rosalind Franklin, and Maurice Wilkins played crucial roles in deciphering the Nucleic acid doub ...
presented their molecular modeling
Molecular modelling encompasses all methods, theoretical and computational, used to model or mimic the behaviour of molecules. The methods are used in the fields of computational chemistry, drug design, computational biology and materials scienc ...
analysis of the DNA X-ray diffraction patterns to suggest that the structure was a double helix.right-handed
In human biology, handedness is an individual's preferential use of one hand, known as the dominant hand, due to and causing it to be stronger, faster or more Fine motor skill, dextrous. The other hand, comparatively often the weaker, less dext ...
spiral, with a shallow, wide minor groove and a narrower, deeper major groove. The A form occurs under non-physiological conditions in partly dehydrated samples of DNA, while in the cell it may be produced in hybrid pairings of DNA and RNA strands, and in enzyme-DNA complexes. Segments of DNA where the bases have been chemically modified by methylation
Methylation, in the chemistry, chemical sciences, is the addition of a methyl group on a substrate (chemistry), substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replac ...
may undergo a larger change in conformation and adopt the Z form. Here, the strands turn about the helical axis in a left-handed spiral, the opposite of the more common B form. These unusual structures can be recognized by specific Z-DNA binding proteins and may be involved in the regulation of transcription.
Alternative DNA chemistry
For many years, exobiologists have proposed the existence of a shadow biosphere
A shadow biosphere is the hypothesis of a hypothetical microbial biosphere of Earth that would use radically different biochemical and molecular processes from that of currently known life. Although life on Earth is relatively well studied, if a s ...
, a postulated microbial biosphere
The biosphere (), also called the ecosphere (), is the worldwide sum of all ecosystems. It can also be termed the zone of life on the Earth. The biosphere (which is technically a spherical shell) is virtually a closed system with regard to mat ...
of Earth that uses radically different biochemical and molecular processes than currently known life. One of the proposals was the existence of lifeforms that use arsenic instead of phosphorus in DNA. A report in 2010 of the possibility in the bacterium
Bacteria (; : bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the ...
GFAJ-1
GFAJ-1 is a strain of rod-shaped bacteria in the family Halomonadaceae. It is an extremophile that was isolated from the hypersaline and alkaline Mono Lake in eastern California by geobiologist Felisa Wolfe-Simon, a NASA research fellow in re ...
was announced,
Quadruplex structures
 At the ends of the linear chromosomes are specialized regions of DNA called
At the ends of the linear chromosomes are specialized regions of DNA called telomere
A telomere (; ) is a region of repetitive nucleotide sequences associated with specialized proteins at the ends of linear chromosomes (see #Sequences, Sequences). Telomeres are a widespread genetic feature most commonly found in eukaryotes. In ...
s. The main function of these regions is to allow the cell to replicate chromosome ends using the enzyme telomerase
Telomerase, also called terminal transferase, is a ribonucleoprotein that adds a species-dependent telomere repeat sequence to the 3' end of telomeres. A telomere is a region of repetitive sequences at each end of the chromosomes of most euka ...
, as the enzymes that normally replicate DNA cannot copy the extreme 3′ ends of chromosomes.DNA repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is cons ...
systems in the cell from treating them as damage to be corrected.human cells
The list of human cell types provides an enumeration and description of the various specialized cells found within the human body, highlighting their distinct functions, characteristics, and contributions to overall physiological processes. Cell ...
, telomeres are usually lengths of single-stranded DNA containing several thousand repeats of a simple TTAGGG sequence.
These guanine-rich sequences may stabilize chromosome ends by forming structures of stacked sets of four-base units, rather than the usual base pairs found in other DNA molecules. Here, four guanine bases, known as a guanine tetrad
In molecular biology, a guanine tetrad (also known as a G-tetrad or G-quartet) is a structure composed of four guanine bases in a square planar array. They most prominently contribute to the structure of G-quadruplexes, where their hydrogen bon ...
, form a flat plate. These flat four-base units then stack on top of each other to form a stable G-quadruplex
In molecular biology, G-quadruplex secondary structures (G4) are formed in nucleic acids by sequences that are rich in guanine. They are helical in shape and contain guanine tetrads that can form from one, two or four strands. The unimolecular ...
structure.chelation
Chelation () is a type of bonding of ions and their molecules to metal ions. It involves the formation or presence of two or more separate coordinate bonds between a polydentate (multiple bonded) ligand and a single central metal atom. These l ...
of a metal ion in the centre of each four-base unit. Other structures can also be formed, with the central set of four bases coming from either a single strand folded around the bases, or several different parallel strands, each contributing one base to the central structure.
In addition to these stacked structures, telomeres also form large loop structures called telomere loops, or T-loops. Here, the single-stranded DNA curls around in a long circle stabilized by telomere-binding proteins. At the very end of the T-loop, the single-stranded telomere DNA is held onto a region of double-stranded DNA by the telomere strand disrupting the double-helical DNA and base pairing to one of the two strands. This triple-stranded structure is called a displacement loop or D-loop
In molecular biology, a displacement loop or D-loop is a DNA structure where the two strands of a double-stranded DNA molecule are separated for a stretch and held apart by a third strand of DNA. An R-loop is similar to a D-loop, but in that cas ...
.
Branched DNA
Branched DNA can form networks containing multiple branches.
In DNA, fraying occurs when non-complementary regions exist at the end of an otherwise complementary double-strand of DNA. However, branched DNA can occur if a third strand of DNA is introduced and contains adjoining regions able to hybridize with the frayed regions of the pre-existing double-strand. Although the simplest example of branched DNA involves only three strands of DNA, complexes involving additional strands and multiple branches are also possible. Branched DNA can be used in nanotechnology
Nanotechnology is the manipulation of matter with at least one dimension sized from 1 to 100 nanometers (nm). At this scale, commonly known as the nanoscale, surface area and quantum mechanical effects become important in describing propertie ...
to construct geometric shapes, see the section on uses in technology below.
Artificial bases
Several artificial nucleobases have been synthesized, and successfully incorporated in the eight-base DNA analogue named Hachimoji DNA. Dubbed S, B, P, and Z, these artificial bases are capable of bonding with each other in a predictable way (S–B and P–Z), maintain the double helix structure of DNA, and be transcribed to RNA. Their existence could be seen as an indication that there is nothing special about the four natural nucleobases that evolved on Earth. On the other hand, DNA is tightly related to RNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA and deoxyrib ...
which does not only act as a transcript of DNA but also performs as molecular machines many tasks in cells. For this purpose it has to fold into a structure. It has been shown that to allow to create all possible structures at least four bases are required for the corresponding RNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA and deoxyrib ...
, while a higher number is also possible but this would be against the natural principle of least effort
The principle of least effort is a broad theory that covers diverse fields from evolutionary biology to webpage design. It postulates that animals, people, and even well-designed machines will naturally choose the path of least resistance or "e ...
.
Acidity
The phosphate groups of DNA give it similar acid
An acid is a molecule or ion capable of either donating a proton (i.e. Hydron, hydrogen cation, H+), known as a Brønsted–Lowry acid–base theory, Brønsted–Lowry acid, or forming a covalent bond with an electron pair, known as a Lewis ...
ic properties to phosphoric acid
Phosphoric acid (orthophosphoric acid, monophosphoric acid or phosphoric(V) acid) is a colorless, odorless phosphorus-containing solid, and inorganic compound with the chemical formula . It is commonly encountered as an 85% aqueous solution, ...
and it can be considered as a strong acid
Acid strength is the tendency of an acid, symbolised by the chemical formula , to dissociate into a hydron (chemistry), proton, , and an anion, . The Dissociation (chemistry), dissociation or ionization of a strong acid in solution is effectivel ...
. It will be fully ionized at a normal cellular pH, releasing proton
A proton is a stable subatomic particle, symbol , Hydron (chemistry), H+, or 1H+ with a positive electric charge of +1 ''e'' (elementary charge). Its mass is slightly less than the mass of a neutron and approximately times the mass of an e ...
s which leave behind negative charges on the phosphate groups. These negative charges protect DNA from breakdown by hydrolysis
Hydrolysis (; ) is any chemical reaction in which a molecule of water breaks one or more chemical bonds. The term is used broadly for substitution reaction, substitution, elimination reaction, elimination, and solvation reactions in which water ...
by repelling nucleophile
In chemistry, a nucleophile is a chemical species that forms bonds by donating an electron pair. All molecules and ions with a free pair of electrons or at least one pi bond can act as nucleophiles. Because nucleophiles donate electrons, they are ...
s which could hydrolyze it.
Macroscopic appearance
Pure DNA extracted from cells forms white, stringy clumps.
Chemical modifications and altered DNA packaging
Base modifications and DNA packaging
Structure of cytosine with and without the 5-methyl group.
Deamination
Deamination is the removal of an amino group from a molecule. Enzymes that catalysis, catalyse this reaction are called deaminases.
In the human body, deamination takes place primarily in the liver; however, it can also occur in the kidney. In s ...
converts 5-methylcytosine into thymine.
The expression of genes is influenced by how the DNA is packaged in chromosomes, in a structure called chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important r ...
. Base modifications can be involved in packaging, with regions that have low or no gene expression usually containing high levels of methylation
Methylation, in the chemistry, chemical sciences, is the addition of a methyl group on a substrate (chemistry), substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replac ...
of cytosine
Cytosine () (symbol C or Cyt) is one of the four nucleotide bases found in DNA and RNA, along with adenine, guanine, and thymine ( uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attac ...
bases. DNA packaging and its influence on gene expression can also occur by covalent modifications of the histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei and in most Archaeal phyla. They act as spools around which DNA winds to create structural units called nucleosomes ...
protein core around which DNA is wrapped in the chromatin structure or else by remodeling carried out by chromatin remodeling complexes (see Chromatin remodeling
Chromatin remodeling is the dynamic modification of chromatin architecture to allow access of condensed genomic DNA to the regulatory transcription machinery proteins, and thereby control gene expression. Such remodeling is principally carried out ...
). There is, further, crosstalk
In electronics, crosstalk (XT) is a phenomenon by which a signal transmitted on one circuit or channel of a transmission system creates an undesired effect in another circuit or channel. Crosstalk is usually caused by undesired capacitive, ...
between DNA methylation and histone modification, so they can coordinately affect chromatin and gene expression.
For one example, cytosine methylation produces 5-methylcytosine
5-Methylcytosine (5mC) is a methylation, methylated form of the DNA base cytosine (C) that regulates gene Transcription (genetics), transcription and takes several other biological roles. When cytosine is methylated, the DNA maintains the same s ...
, which is important for X-inactivation
X-inactivation (also called Lyonization, after English geneticist Mary Lyon) is a process by which one of the copies of the X chromosome is inactivated in therian female mammals. The inactive X chromosome is silenced by being packaged into ...
of chromosomes. The average level of methylation varies between organisms—the worm ''Caenorhabditis elegans
''Caenorhabditis elegans'' () is a free-living transparent nematode about 1 mm in length that lives in temperate soil environments. It is the type species of its genus. The name is a Hybrid word, blend of the Greek ''caeno-'' (recent), ''r ...
'' lacks cytosine methylation, while vertebrate
Vertebrates () are animals with a vertebral column (backbone or spine), and a cranium, or skull. The vertebral column surrounds and protects the spinal cord, while the cranium protects the brain.
The vertebrates make up the subphylum Vertebra ...
s have higher levels, with up to 1% of their DNA containing 5-methylcytosine. Despite the importance of 5-methylcytosine, it can deaminate
Deamination is the removal of an amino group from a molecule. Enzymes that catalyse this reaction are called deaminases.
In the human body, deamination takes place primarily in the liver; however, it can also occur in the kidney. In situations o ...
to leave a thymine base, so methylated cytosines are particularly prone to mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, ...
s. Other base modifications include adenine methylation in bacteria, the presence of 5-hydroxymethylcytosine
5-Hydroxymethylcytosine (5hmC) is a DNA pyrimidine nitrogen base derived from cytosine. It is potentially important in epigenetics, because the hydroxymethyl group on the cytosine can possibly switch a gene on and off. It was first seen in bact ...
in the brain
The brain is an organ (biology), organ that serves as the center of the nervous system in all vertebrate and most invertebrate animals. It consists of nervous tissue and is typically located in the head (cephalization), usually near organs for ...
, and the glycosylation
Glycosylation is the reaction in which a carbohydrate (or ' glycan'), i.e. a glycosyl donor, is attached to a hydroxyl or other functional group of another molecule (a glycosyl acceptor) in order to form a glycoconjugate. In biology (but not ...
of uracil to produce the "J-base" in kinetoplastids
Kinetoplastida (or Kinetoplastea, as a class) is a group of flagellated protists belonging to the phylum Euglenozoa, and characterised by the presence of a distinctive organelle called the kinetoplast (hence the name), a granule containing a la ...
.
Damage
 DNA can be damaged by many sorts of
DNA can be damaged by many sorts of mutagen
In genetics, a mutagen is a physical or chemical agent that permanently changes genetic material, usually DNA, in an organism and thus increases the frequency of mutations above the natural background level. As many mutations can cause cancer in ...
s, which change the DNA sequence
A nucleic acid sequence is a succession of bases within the nucleotides forming alleles within a DNA (using GACT) or RNA (GACU) molecule. This succession is denoted by a series of a set of five different letters that indicate the order of the nu ...
. Mutagens include oxidizing agent
An oxidizing agent (also known as an oxidant, oxidizer, electron recipient, or electron acceptor) is a substance in a redox chemical reaction that gains or " accepts"/"receives" an electron from a (called the , , or ''electron donor''). In ot ...
s, alkylating agents Alkylation is a chemical reaction that entails transfer of an alkyl group. The alkyl group may be transferred as an alkyl carbocation, a free radical, a carbanion, or a carbene (or their equivalents). Alkylating agents are reagents for effectin ...
and also high-energy electromagnetic radiation
In physics, electromagnetic radiation (EMR) is a self-propagating wave of the electromagnetic field that carries momentum and radiant energy through space. It encompasses a broad spectrum, classified by frequency or its inverse, wavelength ...
such as ultraviolet
Ultraviolet radiation, also known as simply UV, is electromagnetic radiation of wavelengths of 10–400 nanometers, shorter than that of visible light, but longer than X-rays. UV radiation is present in sunlight and constitutes about 10% of ...
light and X-ray
An X-ray (also known in many languages as Röntgen radiation) is a form of high-energy electromagnetic radiation with a wavelength shorter than those of ultraviolet rays and longer than those of gamma rays. Roughly, X-rays have a wavelength ran ...
s. The type of DNA damage produced depends on the type of mutagen. For example, UV light can damage DNA by producing thymine dimers, which are cross-links between pyrimidine bases. On the other hand, oxidants such as free radicals
In chemistry, a radical, also known as a free radical, is an atom, molecule, or ion that has at least one unpaired electron, unpaired valence electron.
With some exceptions, these unpaired electrons make radicals highly chemical reaction, chemi ...
or hydrogen peroxide
Hydrogen peroxide is a chemical compound with the formula . In its pure form, it is a very pale blue liquid that is slightly more viscosity, viscous than Properties of water, water. It is used as an oxidizer, bleaching agent, and antiseptic, usua ...
produce multiple forms of damage, including base modifications, particularly of guanosine, and double-strand breaks. A typical human cell contains about 150,000 bases that have suffered oxidative damage. Of these oxidative lesions, the most dangerous are double-strand breaks, as these are difficult to repair and can produce point mutation
A point mutation is a genetic mutation where a single nucleotide base is changed, inserted or deleted from a DNA or RNA sequence of an organism's genome. Point mutations have a variety of effects on the downstream protein product—consequences ...
s, insertions, deletions from the DNA sequence, and chromosomal translocation
In genetics, chromosome translocation is a phenomenon that results in unusual rearrangement of chromosomes. This includes "balanced" and "unbalanced" translocation, with three main types: "reciprocal", "nonreciprocal" and "Robertsonian" transloc ...
s. These mutations can cause cancer
Cancer is a group of diseases involving Cell growth#Disorders, abnormal cell growth with the potential to Invasion (cancer), invade or Metastasis, spread to other parts of the body. These contrast with benign tumors, which do not spread. Po ...
. Because of inherent limits in the DNA repair mechanisms, if humans lived long enough, they would all eventually develop cancer.naturally occurring
A natural product is a natural compound or substance produced by a living organism—that is, found in nature. In the broadest sense, natural products include any substance produced by life. Natural products can also be prepared by chemical ...
, due to normal cellular processes that produce reactive oxygen species, the hydrolytic activities of cellular water, etc., also occur frequently. Although most of these damages are repaired, in any cell some DNA damage may remain despite the action of repair processes. These remaining DNA damages accumulate with age in mammalian postmitotic tissues. This accumulation appears to be an important underlying cause of aging.
Many mutagens fit into the space between two adjacent base pairs, this is called '' intercalation''. Most intercalators are aromatic
In organic chemistry, aromaticity is a chemical property describing the way in which a conjugated system, conjugated ring of unsaturated bonds, lone pairs, or empty orbitals exhibits a stabilization stronger than would be expected from conjugati ...
and planar molecules; examples include ethidium bromide
Ethidium bromide (or homidium bromide, chloride salt homidium chloride) is an intercalating agent commonly used as a fluorescent tag (nucleic acid stain) in molecular biology laboratories for techniques such as agarose gel electrophoresis. It ...
, acridine
Acridine is an organic compound and a nitrogen heterocycle with the formula C13H9N. Acridines are substituted derivatives of the parent ring. It is a planar molecule that is structurally related to anthracene with one of the central CH groups ...
s, daunomycin, and doxorubicin
Doxorubicin, sold under the brand name Adriamycin among others, is a chemotherapy medication used to treat cancer. This includes breast cancer, bladder cancer, Kaposi's sarcoma, lymphoma, and acute lymphocytic leukemia. It is often used toge ...
. For an intercalator to fit between base pairs, the bases must separate, distorting the DNA strands by unwinding of the double helix. This inhibits both transcription and DNA replication, causing toxicity and mutations. As a result, DNA intercalators may be carcinogen
A carcinogen () is any agent that promotes the development of cancer. Carcinogens can include synthetic chemicals, naturally occurring substances, physical agents such as ionizing and non-ionizing radiation, and biologic agents such as viruse ...
s, and in the case of thalidomide, a teratogen. Others such as benzo 'a''yrene diol epoxide and aflatoxin
Aflatoxins are various toxicity, poisonous carcinogens and mutagens that are produced by certain Mold (fungus), molds, especially ''Aspergillus'' species such as ''Aspergillus flavus'' and ''Aspergillus parasiticus''. According to the USDA, "The ...
form DNA adducts that induce errors in replication. Nevertheless, due to their ability to inhibit DNA transcription and replication, other similar toxins are also used in chemotherapy
Chemotherapy (often abbreviated chemo, sometimes CTX and CTx) is the type of cancer treatment that uses one or more anti-cancer drugs (list of chemotherapeutic agents, chemotherapeutic agents or alkylating agents) in a standard chemotherapy re ...
to inhibit rapidly growing cancer
Cancer is a group of diseases involving Cell growth#Disorders, abnormal cell growth with the potential to Invasion (cancer), invade or Metastasis, spread to other parts of the body. These contrast with benign tumors, which do not spread. Po ...
cells.
Biological functions
 DNA usually occurs as linear
DNA usually occurs as linear chromosome
A chromosome is a package of DNA containing part or all of the genetic material of an organism. In most chromosomes, the very long thin DNA fibers are coated with nucleosome-forming packaging proteins; in eukaryotic cells, the most import ...
s in eukaryote
The eukaryotes ( ) constitute the Domain (biology), domain of Eukaryota or Eukarya, organisms whose Cell (biology), cells have a membrane-bound cell nucleus, nucleus. All animals, plants, Fungus, fungi, seaweeds, and many unicellular organisms ...
s, and circular chromosomes in prokaryote
A prokaryote (; less commonly spelled procaryote) is a unicellular organism, single-celled organism whose cell (biology), cell lacks a cell nucleus, nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Ancient Gree ...
s. The set of chromosomes in a cell makes up its genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as ...
; the human genome
The human genome is a complete set of nucleic acid sequences for humans, encoded as the DNA within each of the 23 distinct chromosomes in the cell nucleus. A small DNA molecule is found within individual Mitochondrial DNA, mitochondria. These ar ...
has approximately 3 billion base pairs of DNA arranged into 46 chromosomes.sequence
In mathematics, a sequence is an enumerated collection of objects in which repetitions are allowed and order matters. Like a set, it contains members (also called ''elements'', or ''terms''). The number of elements (possibly infinite) is cal ...
of pieces of DNA called gene
In biology, the word gene has two meanings. The Mendelian gene is a basic unit of heredity. The molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protei ...
s. Transmission
Transmission or transmit may refer to:
Science and technology
* Power transmission
** Electric power transmission
** Transmission (mechanical device), technology that allows controlled application of power
*** Automatic transmission
*** Manual tra ...
of genetic information in genes is achieved via complementary base pairing. For example, in transcription, when a cell uses the information in a gene, the DNA sequence is copied into a complementary RNA sequence through the attraction between the DNA and the correct RNA nucleotides. Usually, this RNA copy is then used to make a matching protein sequence
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthe ...
in a process called translation
Translation is the communication of the semantics, meaning of a #Source and target languages, source-language text by means of an Dynamic and formal equivalence, equivalent #Source and target languages, target-language text. The English la ...
, which depends on the same interaction between RNA nucleotides. In an alternative fashion, a cell may copy its genetic information in a process called DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all life, living organisms, acting as the most essential part of heredity, biolog ...
. The details of these functions are covered in other articles; here the focus is on the interactions between DNA and other molecules that mediate the function of the genome.
Genes and genomes
Genomic DNA is tightly and orderly packed in the process called DNA condensation
DNA condensation refers to the process of compacting DNA molecules ''in vitro'' or ''in vivo''. Mechanistic details of DNA packing are essential for its functioning in the process of gene regulation in living systems. Condensed DNA often has surpr ...
, to fit the small available volumes of the cell. In eukaryotes, DNA is located in the cell nucleus
The cell nucleus (; : nuclei) is a membrane-bound organelle found in eukaryote, eukaryotic cell (biology), cells. Eukaryotic cells usually have a single nucleus, but a few cell types, such as mammalian red blood cells, have #Anucleated_cells, ...
, with small amounts in mitochondria
A mitochondrion () is an organelle found in the cells of most eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is us ...
and chloroplast
A chloroplast () is a type of membrane-bound organelle, organelle known as a plastid that conducts photosynthesis mostly in plant cell, plant and algae, algal cells. Chloroplasts have a high concentration of chlorophyll pigments which captur ...
s. In prokaryotes, the DNA is held within an irregularly shaped body in the cytoplasm called the nucleoid
The nucleoid (meaning '' nucleus-like'') is an irregularly shaped region within the prokaryotic cell that contains all or most of the genetic material. The chromosome of a typical prokaryote is circular, and its length is very large compared to ...
. The genetic information in a genome is held within genes, and the complete set of this information in an organism is called its genotype
The genotype of an organism is its complete set of genetic material. Genotype can also be used to refer to the alleles or variants an individual carries in a particular gene or genetic location. The number of alleles an individual can have in a ...
. A gene is a unit of heredity
Heredity, also called inheritance or biological inheritance, is the passing on of traits from parents to their offspring; either through asexual reproduction or sexual reproduction, the offspring cells or organisms acquire the genetic infor ...
and is a region of DNA that influences a particular characteristic in an organism. Genes contain an open reading frame
In molecular biology, reading frames are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a prokaryotic DNA sequence, where only one of the six possible reading frames ...
that can be transcribed, and regulatory sequence
A regulatory sequence is a segment of a nucleic acid molecule which is capable of increasing or decreasing the expression of specific genes within an organism. Regulation of gene expression is an essential feature of all living organisms and vir ...
s such as promoters and enhancers
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins ( activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcriptio ...
, which control transcription of the open reading frame.
In many species
A species () is often defined as the largest group of organisms in which any two individuals of the appropriate sexes or mating types can produce fertile offspring, typically by sexual reproduction. It is the basic unit of Taxonomy (biology), ...
, only a small fraction of the total sequence of the genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as ...
encodes protein. For example, only about 1.5% of the human genome consists of protein-coding exon
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequence ...
s, with over 50% of human DNA consisting of non-coding repetitive sequences. The reasons for the presence of so much noncoding DNA
Non-coding DNA (ncDNA) sequences are components of an organism's DNA that do not encode protein sequences. Some non-coding DNA is transcribed into functional non-coding RNA molecules (e.g. transfer RNA, microRNA, piRNA, ribosomal RNA, and regu ...
in eukaryotic genomes and the extraordinary differences in genome size
Genome size is the total amount of DNA contained within one copy of a single complete genome. It is typically measured in terms of mass in picograms (trillionths or 10−12 of a gram, abbreviated pg) or less frequently in daltons, or as the tot ...
, or '' C-value'', among species, represent a long-standing puzzle known as the " C-value enigma". However, some DNA sequences that do not code protein may still encode functional non-coding RNA
A non-coding RNA (ncRNA) is a functional RNA molecule that is not Translation (genetics), translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally imp ...
molecules, which are involved in the regulation of gene expression
Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products (protein or RNA). Sophisticated programs of gene expression are wide ...
.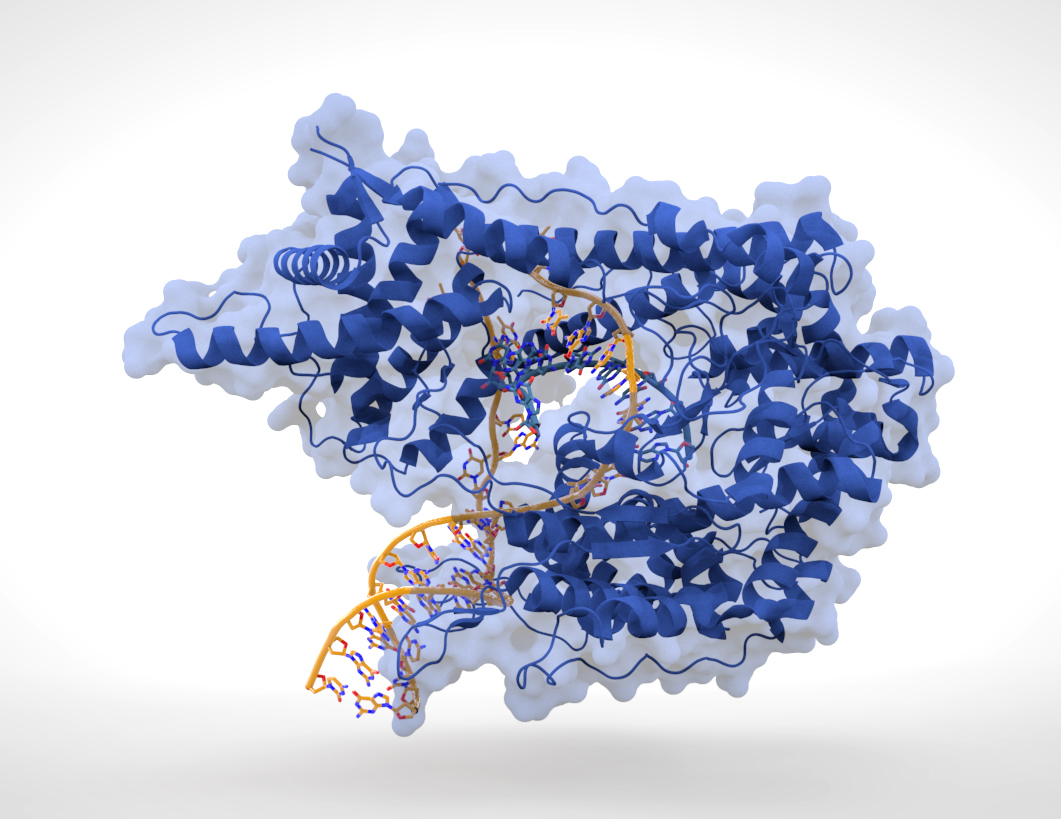 Some noncoding DNA sequences play structural roles in chromosomes.
Some noncoding DNA sequences play structural roles in chromosomes. Telomere
A telomere (; ) is a region of repetitive nucleotide sequences associated with specialized proteins at the ends of linear chromosomes (see #Sequences, Sequences). Telomeres are a widespread genetic feature most commonly found in eukaryotes. In ...
s and centromere
The centromere links a pair of sister chromatids together during cell division. This constricted region of chromosome connects the sister chromatids, creating a short arm (p) and a long arm (q) on the chromatids. During mitosis, spindle fiber ...
s typically contain few genes but are important for the function and stability of chromosomes.pseudogene
Pseudogenes are nonfunctional segments of DNA that resemble functional genes. Pseudogenes can be formed from both protein-coding genes and non-coding genes. In the case of protein-coding genes, most pseudogenes arise as superfluous copies of fun ...
s, which are copies of genes that have been disabled by mutation. These sequences are usually just molecular fossil
A fossil (from Classical Latin , ) is any preserved remains, impression, or trace of any once-living thing from a past geological age. Examples include bones, shells, exoskeletons, stone imprints of animals or microbes, objects preserve ...
s, although they can occasionally serve as raw genetic material
Nucleic acids are large biomolecules that are crucial in all cells and viruses. They are composed of nucleotides, which are the monomer components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main classes of nucleic aci ...
for the creation of new genes through the process of gene duplication
Gene duplication (or chromosomal duplication or gene amplification) is a major mechanism through which new genetic material is generated during molecular evolution. It can be defined as any duplication of a region of DNA that contains a gene ...
and divergence
In vector calculus, divergence is a vector operator that operates on a vector field, producing a scalar field giving the rate that the vector field alters the volume in an infinitesimal neighborhood of each point. (In 2D this "volume" refers to ...
.
Transcription and translation
A gene is a sequence of DNA that contains genetic information and can influence the phenotype
In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology (physical form and structure), its developmental processes, its biochemical and physiological propert ...
of an organism. Within a gene, the sequence of bases along a DNA strand defines a messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
sequence, which then defines one or more protein sequences. The relationship between the nucleotide sequences of genes and the amino-acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 α-amino acids incorporated into proteins. Only these 22 a ...
sequences of proteins is determined by the rules of translation
Translation is the communication of the semantics, meaning of a #Source and target languages, source-language text by means of an Dynamic and formal equivalence, equivalent #Source and target languages, target-language text. The English la ...
, known collectively as the genetic code
Genetic code is a set of rules used by living cell (biology), cells to Translation (biology), translate information encoded within genetic material (DNA or RNA sequences of nucleotide triplets or codons) into proteins. Translation is accomplished ...
. The genetic code consists of three-letter 'words' called ''codons'' formed from a sequence of three nucleotides (e.g. ACT, CAG, TTT).
In transcription, the codons of a gene are copied into messenger RNA by RNA polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that catalyzes the chemical reactions that synthesize RNA from a DNA template.
Using the e ...
. This RNA copy is then decoded by a ribosome
Ribosomes () are molecular machine, macromolecular machines, found within all cell (biology), cells, that perform Translation (biology), biological protein synthesis (messenger RNA translation). Ribosomes link amino acids together in the order s ...
that reads the RNA sequence by base-pairing the messenger RNA to transfer RNA
Transfer ribonucleic acid (tRNA), formerly referred to as soluble ribonucleic acid (sRNA), is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes). In a cell, it provides the physical link between the gene ...
, which carries amino acids. Since there are 4 bases in 3-letter combinations, there are 64 possible codons (43 combinations). These encode the twenty standard amino acids, giving most amino acids more than one possible codon. There are also three 'stop' or 'nonsense' codons signifying the end of the coding region; these are the TAG, TAA, and TGA codons, (UAG, UAA, and UGA on the mRNA).
Replication

Cell division
Cell division is the process by which a parent cell (biology), cell divides into two daughter cells. Cell division usually occurs as part of a larger cell cycle in which the cell grows and replicates its chromosome(s) before dividing. In eukar ...
is essential for an organism to grow, but, when a cell divides, it must replicate the DNA in its genome so that the two daughter cells have the same genetic information as their parent. The double-stranded structure of DNA provides a simple mechanism for DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all life, living organisms, acting as the most essential part of heredity, biolog ...
. Here, the two strands are separated and then each strand's complementary DNA
In genetics, complementary DNA (cDNA) is DNA that was reverse transcribed (via reverse transcriptase) from an RNA (e.g., messenger RNA or microRNA). cDNA exists in both single-stranded and double-stranded forms and in both natural and engin ...
sequence is recreated by an enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
called DNA polymerase
A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create t ...
. This enzyme makes the complementary strand by finding the correct base through complementary base pairing and bonding it onto the original strand. As DNA polymerases can only extend a DNA strand in a 5′ to 3′ direction, different mechanisms are used to copy the antiparallel strands of the double helix. In this way, the base on the old strand dictates which base appears on the new strand, and the cell ends up with a perfect copy of its DNA.
Extracellular nucleic acids
Naked extracellular DNA (eDNA), most of it released by cell death, is nearly ubiquitous in the environment. Its concentration in soil may be as high as 2 μg/L, and its concentration in natural aquatic environments may be as high at 88 μg/L.horizontal gene transfer
Horizontal gene transfer (HGT) or lateral gene transfer (LGT) is the movement of genetic material between organisms other than by the ("vertical") transmission of DNA from parent to offspring (reproduction). HGT is an important factor in the e ...
;biofilm
A biofilm is a Syntrophy, syntrophic Microbial consortium, community of microorganisms in which cell (biology), cells cell adhesion, stick to each other and often also to a surface. These adherent cells become embedded within a slimy ext ...
s of several bacterial species. It may act as a recognition factor to regulate the attachment and dispersal of specific cell types in the biofilm;Cell-free fetal DNA
Cell-free fetal DNA (cffDNA) is fetal DNA that circulates freely in the maternal blood. Maternal blood is sampled by venipuncture. Analysis of cffDNA is a method of non-invasive prenatal diagnosis frequently ordered for pregnant women of advance ...
is found in the blood of the mother, and can be sequenced to determine a great deal of information about the developing fetus.environmental DNA
Environmental DNA or eDNA is DNA that is collected from a variety of environmental samples such as soil, seawater, snow or air, rather than directly sampled from an individual organism. As various organisms interact with the environment, DNA ...
eDNA has seen increased use in the natural sciences as a survey tool for ecology
Ecology () is the natural science of the relationships among living organisms and their Natural environment, environment. Ecology considers organisms at the individual, population, community (ecology), community, ecosystem, and biosphere lev ...
, monitoring the movements and presence of species in water, air, or on land, and assessing an area's biodiversity.
Neutrophil extracellular traps
Neutrophil extracellular traps (NETs) are networks of extracellular fibers, primarily composed of DNA, which allow neutrophils
Neutrophils are a type of phagocytic white blood cell and part of innate immunity. More specifically, they form the most abundant type of granulocytes and make up 40% to 70% of all white blood cells in humans. Their functions vary in different ...
, a type of white blood cell, to kill extracellular pathogens while minimizing damage to the host cells.
Interactions with proteins
All the functions of DNA depend on interactions with proteins. These protein interactions can be non-specific, or the protein can bind specifically to a single DNA sequence. Enzymes can also bind to DNA and of these, the polymerases that copy the DNA base sequence in transcription and DNA replication are particularly important.
DNA-binding proteins
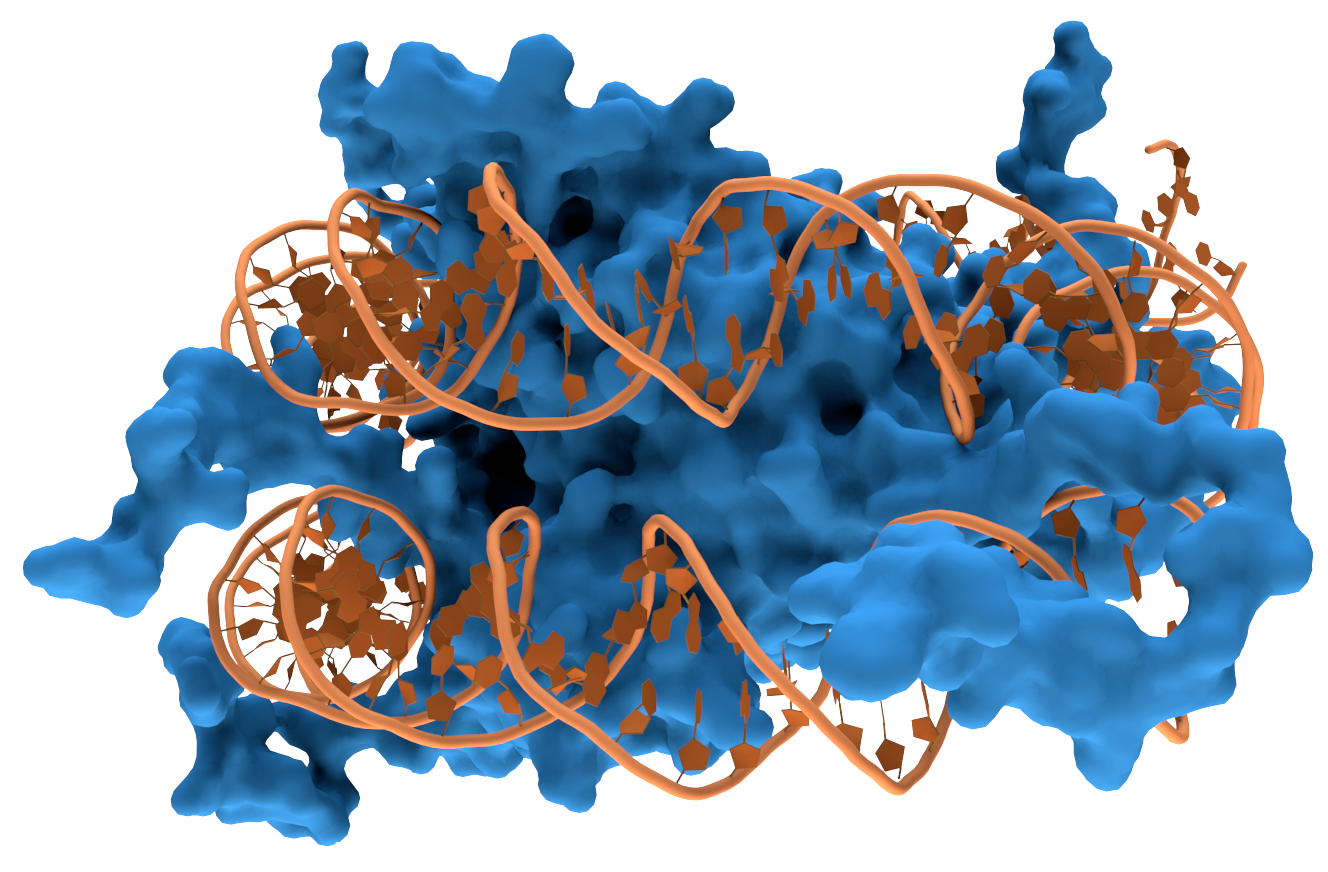 Structural proteins that bind DNA are well-understood examples of non-specific DNA-protein interactions. Within chromosomes, DNA is held in complexes with structural proteins. These proteins organize the DNA into a compact structure called
Structural proteins that bind DNA are well-understood examples of non-specific DNA-protein interactions. Within chromosomes, DNA is held in complexes with structural proteins. These proteins organize the DNA into a compact structure called chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important r ...
. In eukaryotes, this structure involves DNA binding to a complex of small basic proteins called histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei and in most Archaeal phyla. They act as spools around which DNA winds to create structural units called nucleosomes ...
s, while in prokaryotes multiple types of proteins are involved. The histones form a disk-shaped complex called a nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone, histone proteins and resembles thread wrapped around a bobbin, spool. The nucleosome ...
, which contains two complete turns of double-stranded DNA wrapped around its surface. These non-specific interactions are formed through basic residues in the histones, making ionic bond
Ionic bonding is a type of chemical bond
A chemical bond is the association of atoms or ions to form molecules, crystals, and other structures. The bond may result from the electrostatic force between oppositely charged ions as in ionic ...
s to the acidic sugar-phosphate backbone of the DNA, and are thus largely independent of the base sequence. Chemical modifications of these basic amino acid residues include methylation
Methylation, in the chemistry, chemical sciences, is the addition of a methyl group on a substrate (chemistry), substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replac ...
, phosphorylation
In biochemistry, phosphorylation is described as the "transfer of a phosphate group" from a donor to an acceptor. A common phosphorylating agent (phosphate donor) is ATP and a common family of acceptor are alcohols:
:
This equation can be writ ...
, and acetylation
:
In chemistry, acetylation is an organic esterification reaction with acetic acid. It introduces an acetyl group into a chemical compound. Such compounds are termed ''acetate esters'' or simply ''acetates''. Deacetylation is the opposite react ...
. These chemical changes alter the strength of the interaction between the DNA and the histones, making the DNA more or less accessible to transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription (genetics), transcription of genetics, genetic information from DNA to messenger RNA, by binding t ...
s and changing the rate of transcription. Other non-specific DNA-binding proteins in chromatin include the high-mobility group proteins, which bind to bent or distorted DNA. These proteins are important in bending arrays of nucleosomes and arranging them into the larger structures that make up chromosomes.
A distinct group of DNA-binding proteins is the DNA-binding proteins that specifically bind single-stranded DNA. In humans, replication protein A
Protein A is a 42 kDa surface protein originally found in the cell wall of the bacteria ''Staphylococcus aureus''. It is encoded by the ''spa'' gene and its regulation is controlled by DNA topology, cellular osmolarity, and a two-component syst ...
is the best-understood member of this family and is used in processes where the double helix is separated, including DNA replication, recombination, and DNA repair. These binding proteins seem to stabilize single-stranded DNA and protect it from forming stem-loop
Stem-loops are nucleic acid Biomolecular structure, secondary structural elements which form via intramolecular base pairing in single-stranded DNA or RNA. They are also referred to as hairpins or hairpin loops. A stem-loop occurs when two regi ...
s or being degraded by nuclease
In biochemistry, a nuclease (also archaically known as nucleodepolymerase or polynucleotidase) is an enzyme capable of cleaving the phosphodiester bonds that link nucleotides together to form nucleic acids. Nucleases variously affect single and ...
s.
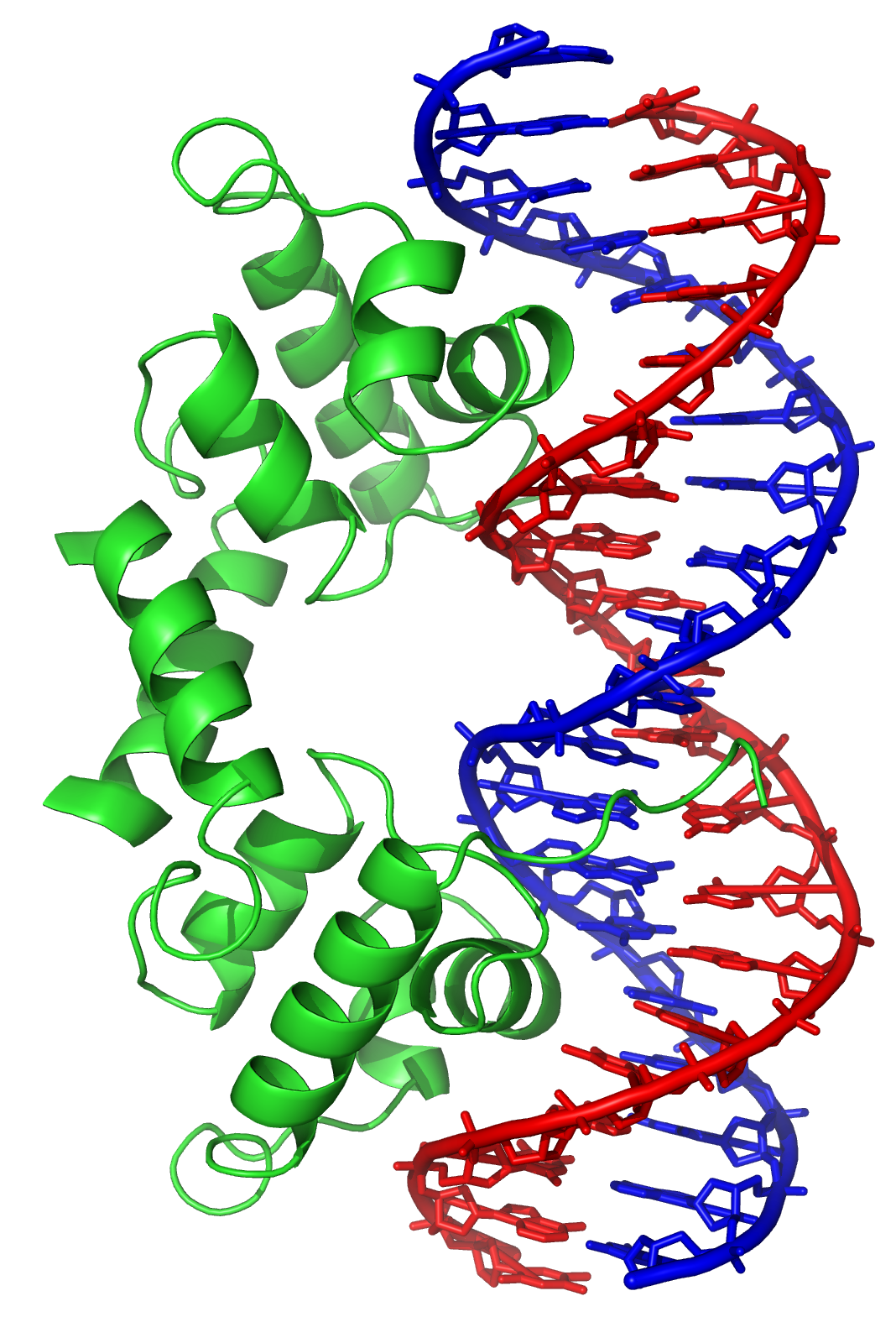 In contrast, other proteins have evolved to bind to particular DNA sequences. The most intensively studied of these are the various
In contrast, other proteins have evolved to bind to particular DNA sequences. The most intensively studied of these are the various transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription (genetics), transcription of genetics, genetic information from DNA to messenger RNA, by binding t ...
s, which are proteins that regulate transcription. Each transcription factor binds to one particular set of DNA sequences and activates or inhibits the transcription of genes that have these sequences close to their promoters. The transcription factors do this in two ways. Firstly, they can bind the RNA polymerase responsible for transcription, either directly or through other mediator proteins; this locates the polymerase at the promoter and allows it to begin transcription. Alternatively, transcription factors can bind enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
s that modify the histones at the promoter. This changes the accessibility of the DNA template to the polymerase.
As these DNA targets can occur throughout an organism's genome, changes in the activity of one type of transcription factor can affect thousands of genes. Consequently, these proteins are often the targets of the signal transduction
Signal transduction is the process by which a chemical or physical signal is transmitted through a cell as a biochemical cascade, series of molecular events. Proteins responsible for detecting stimuli are generally termed receptor (biology), rece ...
processes that control responses to environmental changes or cellular differentiation
Cellular differentiation is the process in which a stem cell changes from one type to a differentiated one. Usually, the cell changes to a more specialized type. Differentiation happens multiple times during the development of a multicellula ...
and development. The specificity of these transcription factors' interactions with DNA come from the proteins making multiple contacts to the edges of the DNA bases, allowing them to "read" the DNA sequence. Most of these base-interactions are made in the major groove, where the bases are most accessible.
DNA-modifying enzymes
Nucleases and ligases

Nuclease
In biochemistry, a nuclease (also archaically known as nucleodepolymerase or polynucleotidase) is an enzyme capable of cleaving the phosphodiester bonds that link nucleotides together to form nucleic acids. Nucleases variously affect single and ...
s are enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
s that cut DNA strands by catalyzing the hydrolysis
Hydrolysis (; ) is any chemical reaction in which a molecule of water breaks one or more chemical bonds. The term is used broadly for substitution reaction, substitution, elimination reaction, elimination, and solvation reactions in which water ...
of the phosphodiester bond
In chemistry, a phosphodiester bond occurs when exactly two of the hydroxyl groups () in phosphoric acid react with hydroxyl groups on other molecules to form two ester bonds. The "bond" involves this linkage . Discussion of phosphodiesters is d ...
s. Nucleases that hydrolyse nucleotides from the ends of DNA strands are called exonuclease
Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is th ...
s, while endonuclease
In molecular biology, endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain (namely DNA or RNA). Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (with regard to sequence), while man ...
s cut within strands. The most frequently used nucleases in molecular biology
Molecular biology is a branch of biology that seeks to understand the molecule, molecular basis of biological activity in and between Cell (biology), cells, including biomolecule, biomolecular synthesis, modification, mechanisms, and interactio ...
are the restriction endonucleases
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class o ...
, which cut DNA at specific sequences. For instance, the EcoRV enzyme shown to the left recognizes the 6-base sequence 5′-GATATC-3′ and makes a cut at the horizontal line. In nature, these enzymes protect bacteria
Bacteria (; : bacterium) are ubiquitous, mostly free-living organisms often consisting of one Cell (biology), biological cell. They constitute a large domain (biology), domain of Prokaryote, prokaryotic microorganisms. Typically a few micr ...
against phage
A bacteriophage (), also known informally as a phage (), is a virus that infects and replicates within bacteria. The term is derived . Bacteriophages are composed of proteins that encapsulate a DNA or RNA genome, and may have structures tha ...
infection by digesting the phage DNA when it enters the bacterial cell, acting as part of the restriction modification system
The restriction modification system (RM system) is found in bacteria and archaea, and provides a defense against foreign DNA, such as that borne by bacteriophages.
Bacteria have restriction enzymes, also called restriction endonucleases, which ...
. In technology, these sequence-specific nucleases are used in molecular cloning
Molecular cloning is a set of experimental methods in molecular biology that are used to assemble recombinant DNA molecules and to direct their DNA replication, replication within Host (biology), host organisms. The use of the word ''cloning'' re ...
and DNA fingerprinting
DNA profiling (also called DNA fingerprinting and genetic fingerprinting) is the process of determining an individual's deoxyribonucleic acid (DNA) characteristics. DNA analysis intended to identify a species, rather than an individual, is cal ...
.
Enzymes called DNA ligase
DNA ligase is a type of enzyme that facilitates the joining of DNA strands together by catalyzing the formation of a phosphodiester bond. It plays a role in repairing single-strand breaks in duplex DNA in living organisms, but some forms (such ...
s can rejoin cut or broken DNA strands.lagging strand
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms, acting as the most essential part of biological inheritance ...
DNA replication, as they join the short segments of DNA produced at the replication fork
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms, acting as the most essential part of biological inheritanc ...
into a complete copy of the DNA template. They are also used in DNA repair
DNA repair is a collection of processes by which a cell (biology), cell identifies and corrects damage to the DNA molecules that encode its genome. A weakened capacity for DNA repair is a risk factor for the development of cancer. DNA is cons ...
and genetic recombination
Genetic recombination (also known as genetic reshuffling) is the exchange of genetic material between different organisms which leads to production of offspring with combinations of traits that differ from those found in either parent. In eukaryot ...
.
Topoisomerases and helicases
Topoisomerase
DNA topoisomerases (or topoisomerases) are enzymes that catalyze changes in the topological state of DNA, interconverting relaxed and supercoiled forms, linked (catenated) and unlinked species, and knotted and unknotted DNA. Topological issues in ...
s are enzymes with both nuclease and ligase activity. These proteins change the amount of supercoiling in DNA. Some of these enzymes work by cutting the DNA helix and allowing one section to rotate, thereby reducing its level of supercoiling; the enzyme then seals the DNA break.Helicase
Helicases are a class of enzymes that are vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic double helix, separating the two hybridized ...
s are proteins that are a type of molecular motor
Molecular motors are natural (biological) or artificial molecular machines that are the essential agents of movement in living organisms. In general terms, a motor is a device that consumes energy in one form and converts it into motion or mech ...
. They use the chemical energy in nucleoside triphosphate
A nucleoside triphosphate is a nucleoside containing a nitrogenous base bound to a 5-carbon sugar (either ribose or deoxyribose), with three phosphate groups bound to the sugar. They are the molecular precursors of both DNA and RNA, which are chai ...
s, predominantly adenosine triphosphate
Adenosine triphosphate (ATP) is a nucleoside triphosphate that provides energy to drive and support many processes in living cell (biology), cells, such as muscle contraction, nerve impulse propagation, and chemical synthesis. Found in all known ...
(ATP), to break hydrogen bonds between bases and unwind the DNA double helix into single strands. These enzymes are essential for most processes where enzymes need to access the DNA bases.
Polymerases
Polymerase
In biochemistry, a polymerase is an enzyme (Enzyme Commission number, EC 2.7.7.6/7/19/48/49) that synthesizes long chains of polymers or nucleic acids. DNA polymerase and RNA polymerase are used to assemble DNA and RNA molecules, respectively, by ...
s are enzyme
An enzyme () is a protein that acts as a biological catalyst by accelerating chemical reactions. The molecules upon which enzymes may act are called substrate (chemistry), substrates, and the enzyme converts the substrates into different mol ...
s that synthesize polynucleotide chains from nucleoside triphosphate
A nucleoside triphosphate is a nucleoside containing a nitrogenous base bound to a 5-carbon sugar (either ribose or deoxyribose), with three phosphate groups bound to the sugar. They are the molecular precursors of both DNA and RNA, which are chai ...
s. The sequence of their products is created based on existing polynucleotide chains—which are called ''templates''. These enzymes function by repeatedly adding a nucleotide to the 3′ hydroxyl
In chemistry, a hydroxy or hydroxyl group is a functional group with the chemical formula and composed of one oxygen atom covalently bonded to one hydrogen atom. In organic chemistry, alcohols and carboxylic acids contain one or more hydroxy ...
group at the end of the growing polynucleotide chain. As a consequence, all polymerases work in a 5′ to 3′ direction.active site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate, the ''binding s ...
of these enzymes, the incoming nucleoside triphosphate base-pairs to the template: this allows polymerases to accurately synthesize the complementary strand of their template. Polymerases are classified according to the type of template that they use.
In DNA replication, DNA-dependent DNA polymerase
A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create t ...
s make copies of DNA polynucleotide chains. To preserve biological information, it is essential that the sequence of bases in each copy are precisely complementary to the sequence of bases in the template strand. Many DNA polymerases have a proofreading
Proofreading is a phase in the process of publishing where galley proofs are compared against the original manuscripts or graphic artworks, to identify transcription errors in the typesetting process. In the past, proofreaders would place corr ...
activity. Here, the polymerase recognizes the occasional mistakes in the synthesis reaction by the lack of base pairing between the mismatched nucleotides. If a mismatch is detected, a 3′ to 5′ exonuclease
Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is th ...
activity is activated and the incorrect base removed. In most organisms, DNA polymerases function in a large complex called the replisome that contains multiple accessory subunits, such as the DNA clamp
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
or helicase
Helicases are a class of enzymes that are vital to all organisms. Their main function is to unpack an organism's genetic material. Helicases are motor proteins that move directionally along a nucleic double helix, separating the two hybridized ...
s.
RNA-dependent DNA polymerases are a specialized class of polymerases that copy the sequence of an RNA strand into DNA. They include reverse transcriptase
A reverse transcriptase (RT) is an enzyme used to convert RNA genome to DNA, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobi ...
, which is a viral enzyme involved in the infection of cells by retrovirus
A retrovirus is a type of virus that inserts a DNA copy of its RNA genome into the DNA of a host cell that it invades, thus changing the genome of that cell. After invading a host cell's cytoplasm, the virus uses its own reverse transcriptase e ...
es, and telomerase
Telomerase, also called terminal transferase, is a ribonucleoprotein that adds a species-dependent telomere repeat sequence to the 3' end of telomeres. A telomere is a region of repetitive sequences at each end of the chromosomes of most euka ...
, which is required for the replication of telomeres.telomeres
A telomere (; ) is a region of repetitive nucleotide sequences associated with specialized proteins at the ends of linear chromosomes (see Sequences). Telomeres are a widespread genetic feature most commonly found in eukaryotes. In most, if not ...
at the ends of chromosomes. Telomeres prevent fusion of the ends of neighboring chromosomes and protect chromosome ends from damage.RNA polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that catalyzes the chemical reactions that synthesize RNA from a DNA template.
Using the e ...
that copies the sequence of a DNA strand into RNA. To begin transcribing a gene, the RNA polymerase binds to a sequence of DNA called a promoter and separates the DNA strands. It then copies the gene sequence into a messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
transcript until it reaches a region of DNA called the terminator, where it halts and detaches from the DNA. As with human DNA-dependent DNA polymerases, RNA polymerase II
RNA polymerase II (RNAP II and Pol II) is a Protein complex, multiprotein complex that Transcription (biology), transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNA pol ...
, the enzyme that transcribes most of the genes in the human genome, operates as part of a large protein complex
A protein complex or multiprotein complex is a group of two or more associated polypeptide chains. Protein complexes are distinct from multidomain enzymes, in which multiple active site, catalytic domains are found in a single polypeptide chain.
...
with multiple regulatory and accessory subunits.
Genetic recombination
Structure of the
Holliday junction
A Holliday junction is a branched nucleic acid structure that contains four double-stranded arms joined. These arms may adopt one of several conformations depending on buffer salt concentrations and the sequence of nucleobases closest to the j ...
intermediate in
genetic recombination
Genetic recombination (also known as genetic reshuffling) is the exchange of genetic material between different organisms which leads to production of offspring with combinations of traits that differ from those found in either parent. In eukaryot ...
. The four separate DNA strands are coloured red, blue, green and yellow.
 A DNA helix usually does not interact with other segments of DNA, and in human cells, the different chromosomes even occupy separate areas in the nucleus called "
A DNA helix usually does not interact with other segments of DNA, and in human cells, the different chromosomes even occupy separate areas in the nucleus called "chromosome territories
In cell biology, chromosome territories are regions of the nucleus preferentially occupied by particular chromosomes.
Interphase chromosomes are long DNA strands that are extensively folded, and are often described as appearing like a bowl of s ...
". This physical separation of different chromosomes is important for the ability of DNA to function as a stable repository for information, as one of the few times chromosomes interact is in chromosomal crossover
Chromosomal crossover, or crossing over, is the exchange of genetic material during sexual reproduction between two homologous chromosomes' sister chromatids, non-sister chromatids that results in recombinant chromosomes. It is one of the fina ...
which occurs during sexual reproduction
Sexual reproduction is a type of reproduction that involves a complex life cycle in which a gamete ( haploid reproductive cells, such as a sperm or egg cell) with a single set of chromosomes combines with another gamete to produce a zygote tha ...
, when genetic recombination
Genetic recombination (also known as genetic reshuffling) is the exchange of genetic material between different organisms which leads to production of offspring with combinations of traits that differ from those found in either parent. In eukaryot ...
occurs. Chromosomal crossover is when two DNA helices break, swap a section and then rejoin.
Recombination allows chromosomes to exchange genetic information and produces new combinations of genes, which increases the efficiency of natural selection
Natural selection is the differential survival and reproduction of individuals due to differences in phenotype. It is a key mechanism of evolution, the change in the Heredity, heritable traits characteristic of a population over generation ...
and can be important in the rapid evolution of new proteins. Genetic recombination can also be involved in DNA repair, particularly in the cell's response to double-strand breaks.
The most common form of chromosomal crossover is homologous recombination
Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids (usually DNA as in Cell (biology), cellular organi ...
, where the two chromosomes involved share very similar sequences. Non-homologous recombination
Illegitimate recombination, or nonhomologous recombination, is the process by which two unrelated double stranded segments of DNA are joined. This insertion of genetic material which is not meant to be adjacent tends to lead to genes being broken c ...
can be damaging to cells, as it can produce chromosomal translocation
In genetics, chromosome translocation is a phenomenon that results in unusual rearrangement of chromosomes. This includes "balanced" and "unbalanced" translocation, with three main types: "reciprocal", "nonreciprocal" and "Robertsonian" transloc ...
s and genetic abnormalities. The recombination reaction is catalyzed by enzymes known as recombinase
Recombinases are genetic recombination enzymes.
Site specific recombinases
DNA recombinases are widely used in multicellular organisms to manipulate the structure of genomes, and to control gene expression. These enzymes, derived from bacteria ( ...
s, such as RAD51
DNA repair protein RAD51 homolog 1 is a protein encoded by the gene ''RAD51''. The enzyme encoded by this gene is a member of the RAD51 protein family which assists in repair of DNA double strand breaks. RAD51 family members are homologous to t ...
. The first step in recombination is a double-stranded break caused by either an endonuclease
In molecular biology, endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain (namely DNA or RNA). Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (with regard to sequence), while man ...
or damage to the DNA. A series of steps catalyzed in part by the recombinase then leads to joining of the two helices by at least one Holliday junction
A Holliday junction is a branched nucleic acid structure that contains four double-stranded arms joined. These arms may adopt one of several conformations depending on buffer salt concentrations and the sequence of nucleobases closest to the j ...
, in which a segment of a single strand in each helix is annealed to the complementary strand in the other helix. The Holliday junction is a tetrahedral junction structure that can be moved along the pair of chromosomes, swapping one strand for another. The recombination reaction is then halted by cleavage of the junction and re-ligation of the released DNA. Only strands of like polarity exchange DNA during recombination. There are two types of cleavage: east-west cleavage and north–south cleavage. The north–south cleavage nicks both strands of DNA, while the east–west cleavage has one strand of DNA intact. The formation of a Holliday junction during recombination makes it possible for genetic diversity, genes to exchange on chromosomes, and expression of wild-type viral genomes.
Evolution
DNA contains the genetic information that allows all forms of life to function, grow and reproduce. However, it is unclear how long in the 4-billion-year history of life
The history of life on Earth traces the processes by which living and extinct organisms evolved, from the earliest emergence of life to the present day. Earth formed about 4.5 billion years ago (abbreviated as ''Ga'', for '' gigaannum'') and ...
DNA has performed this function, as it has been proposed that the earliest forms of life may have used RNA as their genetic material.cell metabolism
''Cell Metabolism'' is a monthly peer-reviewed scientific journal covering physiology, with an emphasis on understanding the molecular basis of how the body self-regulates in the face of change, and how disturbances in these balances can lead to di ...
as it can both transmit genetic information and carry out catalysis
Catalysis () is the increase in rate of a chemical reaction due to an added substance known as a catalyst (). Catalysts are not consumed by the reaction and remain unchanged after it. If the reaction is rapid and the catalyst recycles quick ...
as part of ribozyme
Ribozymes (ribonucleic acid enzymes) are RNA molecules that have the ability to Catalysis, catalyze specific biochemical reactions, including RNA splicing in gene expression, similar to the action of protein enzymes. The 1982 discovery of ribozy ...
s. This ancient RNA world
The RNA world is a hypothetical stage in the evolutionary history of life on Earth in which self-replicating RNA molecules proliferated before the evolution of DNA and proteins. The term also refers to the hypothesis that posits the existence ...
where nucleic acid would have been used for both catalysis and genetics may have influenced the evolution
Evolution is the change in the heritable Phenotypic trait, characteristics of biological populations over successive generations. It occurs when evolutionary processes such as natural selection and genetic drift act on genetic variation, re ...
of the current genetic code based on four nucleotide bases. This would occur, since the number of different bases in such an organism is a trade-off between a small number of bases increasing replication accuracy and a large number of bases increasing the catalytic efficiency of ribozymes. However, there is no direct evidence of ancient genetic systems, as recovery of DNA from most fossils is impossible because DNA survives in the environment for less than one million years, and slowly degrades into short fragments in solution. Claims for older DNA have been made, most notably a report of the isolation of a viable bacterium from a salt crystal 250 million years old, but these claims are controversial.
Building blocks of DNA (adenine
Adenine (, ) (nucleoside#List of nucleosides and corresponding nucleobases, symbol A or Ade) is a purine nucleotide base that is found in DNA, RNA, and Adenosine triphosphate, ATP. Usually a white crystalline subtance. The shape of adenine is ...
, guanine
Guanine () (symbol G or Gua) is one of the four main nucleotide bases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine ( uracil in RNA). In DNA, guanine is paired with cytosine. The guanine nucleoside ...
, and related organic molecules
Some chemical authorities define an organic compound as a chemical compound that contains a carbon–hydrogen or carbon–carbon bond; others consider an organic compound to be any chemical compound that contains carbon. For example, carbon-cont ...
) may have been formed extraterrestrially in outer space.RNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA and deoxyrib ...
organic compound
Some chemical authorities define an organic compound as a chemical compound that contains a carbon–hydrogen or carbon–carbon bond; others consider an organic compound to be any chemical compound that contains carbon. For example, carbon-co ...
s of life
Life, also known as biota, refers to matter that has biological processes, such as Cell signaling, signaling and self-sustaining processes. It is defined descriptively by the capacity for homeostasis, Structure#Biological, organisation, met ...
, including uracil
Uracil () (nucleoside#List of nucleosides and corresponding nucleobases, symbol U or Ura) is one of the four nucleotide bases in the nucleic acid RNA. The others are adenine (A), cytosine (C), and guanine (G). In RNA, uracil binds to adenine via ...
, cytosine
Cytosine () (symbol C or Cyt) is one of the four nucleotide bases found in DNA and RNA, along with adenine, guanine, and thymine ( uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attac ...
, and thymine
Thymine () (symbol T or Thy) is one of the four nucleotide bases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The others are adenine, guanine, and cytosine. Thymine is also known as 5-methyluracil, a pyrimidine ...
, have also been formed in the laboratory under conditions mimicking those found in outer space, using starting chemicals, such as pyrimidine
Pyrimidine (; ) is an aromatic, heterocyclic, organic compound similar to pyridine (). One of the three diazines (six-membered heterocyclics with two nitrogen atoms in the ring), it has nitrogen atoms at positions 1 and 3 in the ring. The oth ...
, found in meteorite
A meteorite is a rock (geology), rock that originated in outer space and has fallen to the surface of a planet or Natural satellite, moon. When the original object enters the atmosphere, various factors such as friction, pressure, and chemical ...
s. Pyrimidine, like polycyclic aromatic hydrocarbons
A Polycyclic aromatic hydrocarbon (PAH) is any member of a class of organic compounds that is composed of multiple fused aromatic rings. Most are produced by the incomplete combustion of organic matter— by engine exhaust fumes, tobacco, incin ...
(PAHs), the most carbon-rich chemical found in the universe
The universe is all of space and time and their contents. It comprises all of existence, any fundamental interaction, physical process and physical constant, and therefore all forms of matter and energy, and the structures they form, from s ...
, may have been formed in red giant
A red giant is a luminous giant star of low or intermediate mass (roughly 0.3–8 solar masses ()) in a late phase of stellar evolution. The stellar atmosphere, outer atmosphere is inflated and tenuous, making the radius large and the surface t ...
s or in interstellar cosmic dust
Cosmic dustalso called extraterrestrial dust, space dust, or star dustis dust that occurs in outer space or has fallen onto Earth. Most cosmic dust particles measure between a few molecules and , such as micrometeoroids (30 μm). Cosmic dust can ...
and gas clouds.Ancient DNA
Ancient DNA (aDNA) is DNA isolated from ancient sources (typically Biological specimen, specimens, but also environmental DNA). Due to degradation processes (including Crosslinking of DNA, cross-linking, deamination and DNA fragmentation, fragme ...
has been recovered from ancient organisms at a timescale where genome evolution can be directly observed, including from extinct organisms up to millions of years old, such as the woolly mammoth
The woolly mammoth (''Mammuthus primigenius'') is an extinct species of mammoth that lived from the Middle Pleistocene until its extinction in the Holocene epoch. It was one of the last in a line of mammoth species, beginning with the African ...
.
Uses in technology
Genetic engineering
Methods have been developed to purify DNA from organisms, such as phenol-chloroform extraction, and to manipulate it in the laboratory, such as restriction digest
In molecular biology, a restriction digest is a procedure used to prepare DNA for analysis or other processing. It is sometimes termed ''DNA fragmentation'', though this term is used for other procedures as well. In a restriction digest, DNA mol ...
s and the polymerase chain reaction
The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA (or a part of it) sufficiently to enable detailed st ...
. Modern biology
Biology is the scientific study of life and living organisms. It is a broad natural science that encompasses a wide range of fields and unifying principles that explain the structure, function, growth, History of life, origin, evolution, and ...
and biochemistry
Biochemistry, or biological chemistry, is the study of chemical processes within and relating to living organisms. A sub-discipline of both chemistry and biology, biochemistry may be divided into three fields: structural biology, enzymology, a ...
make intensive use of these techniques in recombinant DNA technology. Recombinant DNA
Recombinant DNA (rDNA) molecules are DNA molecules formed by laboratory methods of genetic recombination (such as molecular cloning) that bring together genetic material from multiple sources, creating sequences that would not otherwise be fo ...
is a man-made DNA sequence that has been assembled from other DNA sequences. They can be transformed into organisms in the form of plasmid
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria and ...
s or in the appropriate format, by using a viral vector
A viral vector is a modified virus designed to gene delivery, deliver genetic material into cell (biology), cells. This process can be performed inside an organism or in cell culture. Viral vectors have widespread applications in basic research, ...
. The genetically modified
Genetic engineering, also called genetic modification or genetic manipulation, is the modification and manipulation of an organism's genes using technology. It is a set of technologies used to change the genetic makeup of cells, including th ...
organisms produced can be used to produce products such as recombinant protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
s, used in medical research
Medical research (or biomedical research), also known as health research, refers to the process of using scientific methods with the aim to produce knowledge about human diseases, the prevention and treatment of illness, and the promotion of ...
, or be grown in agriculture
Agriculture encompasses crop and livestock production, aquaculture, and forestry for food and non-food products. Agriculture was a key factor in the rise of sedentary human civilization, whereby farming of domesticated species created ...
.
DNA profiling
Forensic scientists
Forensic science combines principles of law and science to investigate criminal activity. Through crime scene investigations and laboratory analysis, forensic scientists are able to link suspects to evidence. An example is determining the time and ...
can use DNA in blood
Blood is a body fluid in the circulatory system of humans and other vertebrates that delivers necessary substances such as nutrients and oxygen to the cells, and transports metabolic waste products away from those same cells.
Blood is com ...
, semen
Semen, also known as seminal fluid, is a bodily fluid that contains spermatozoon, spermatozoa which is secreted by the male gonads (sexual glands) and other sexual organs of male or hermaphrodite, hermaphroditic animals. In humans and placen ...
, skin
Skin is the layer of usually soft, flexible outer tissue covering the body of a vertebrate animal, with three main functions: protection, regulation, and sensation.
Other animal coverings, such as the arthropod exoskeleton, have different ...
, saliva
Saliva (commonly referred as spit or drool) is an extracellular fluid produced and secreted by salivary glands in the mouth. In humans, saliva is around 99% water, plus electrolytes, mucus, white blood cells, epithelial cells (from which ...
or hair
Hair is a protein filament that grows from follicles found in the dermis. Hair is one of the defining characteristics of mammals.
The human body, apart from areas of glabrous skin, is covered in follicles which produce thick terminal and ...
found at a crime scene
A crime scene is any location that may be associated with a committed crime. Crime scenes contain physical evidence that is pertinent to a criminal investigation. This evidence is collected by crime scene investigators (CSI) and law enforcement. ...
to identify a matching DNA of an individual, such as a perpetrator. This process is formally termed DNA profiling
DNA profiling (also called DNA fingerprinting and genetic fingerprinting) is the process of determining an individual's deoxyribonucleic acid (DNA) characteristics. DNA analysis intended to identify a species, rather than an individual, is cal ...
, also called ''DNA fingerprinting''. In DNA profiling, the lengths of variable sections of repetitive DNA, such as short tandem repeat
A microsatellite is a tract of repetitive DNA in which certain DNA motifs (ranging in length from one to six or more base pairs) are repeated, typically 5–50 times. Microsatellites occur at thousands of locations within an organism's genome. T ...
s and minisatellite
In genetics, a minisatellite is a tract of repetitive DNA in which certain DNA motifs (ranging in length from 10–60 base pairs) are typically repeated two to several hundred times. Minisatellites occur at more than 1,000 locations in the huma ...
s, are compared between people. This method is usually an extremely reliable technique for identifying a matching DNA. However, identification can be complicated if the scene is contaminated with DNA from several people. DNA profiling was developed in 1984 by British geneticist Sir Alec Jeffreys
Sir Alec John Jeffreys, (born 9 January 1950) is a British geneticist known for developing techniques for genetic fingerprinting and DNA profiling which are now used worldwide in forensic science to assist police detective work and to resolve ...
, and first used in forensic science to convict Colin Pitchfork in the 1988 Enderby murders case.
The development of forensic science and the ability to now obtain genetic matching on minute samples of blood, skin, saliva, or hair has led to re-examining many cases. Evidence can now be uncovered that was scientifically impossible at the time of the original examination. Combined with the removal of the double jeopardy
In jurisprudence, double jeopardy is a procedural defence (primarily in common law jurisdictions) that prevents an accused person from being tried again on the same (or similar) charges following an acquittal or conviction and in rare cases ...
law in some places, this can allow cases to be reopened where prior trials have failed to produce sufficient evidence to convince a jury. People charged with serious crimes may be required to provide a sample of DNA for matching purposes. The most obvious defense to DNA matches obtained forensically is to claim that cross-contamination of evidence has occurred. This has resulted in meticulous strict handling procedures with new cases of serious crime.
DNA profiling is also used successfully to positively identify victims of mass casualty incidents, bodies or body parts in serious accidents, and individual victims in mass war graves, via matching to family members.
DNA profiling is also used in DNA paternity testing
DNA paternity testing uses DNA profiling, DNA profiles to determine whether an individual is the biology, biological parent of another individual. Paternity testing can be essential when the rights and duties of the father are in issue, and a ch ...
to determine if someone is the biological parent or grandparent of a child with the probability of parentage is typically 99.99% when the alleged parent is biologically related to the child. Normal DNA sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, thymine, cytosine, and guanine. The ...
methods happen after birth, but there are new methods to test paternity while a mother is still pregnant.
DNA enzymes or catalytic DNA
Deoxyribozyme
Deoxyribozymes, also called DNA enzymes, DNAzymes, or catalytic DNA, are DNA oligonucleotides that are capable of performing a specific chemical reaction, often but not always catalytic. This is similar to the action of other biological enzymes, s ...
s, also called DNAzymes or catalytic DNA, were first discovered in 1994.in vitro
''In vitro'' (meaning ''in glass'', or ''in the glass'') Research, studies are performed with Cell (biology), cells or biological molecules outside their normal biological context. Colloquially called "test-tube experiments", these studies in ...
selection or systematic evolution of ligands by exponential enrichment (SELEX). DNAzymes catalyze variety of chemical reactions including RNA-DNA cleavage, RNA-DNA ligation, amino acids phosphorylation-dephosphorylation, carbon-carbon bond formation, etc. DNAzymes can enhance catalytic rate of chemical reactions up to 100,000,000,000-fold over the uncatalyzed reaction. The most extensively studied class of DNAzymes is RNA-cleaving types which have been used to detect different metal ions and designing therapeutic agents. Several metal-specific DNAzymes have been reported including the GR-5 DNAzyme (lead-specific),
Bioinformatics
Bioinformatics
Bioinformatics () is an interdisciplinary field of science that develops methods and Bioinformatics software, software tools for understanding biological data, especially when the data sets are large and complex. Bioinformatics uses biology, ...
involves the development of techniques to store, data mine, search and manipulate biological data, including DNA nucleic acid sequence
A nucleic acid sequence is a succession of Nucleobase, bases within the nucleotides forming alleles within a DNA (using GACT) or RNA (GACU) molecule. This succession is denoted by a series of a set of five different letters that indicate the orde ...
data. These have led to widely applied advances in computer science
Computer science is the study of computation, information, and automation. Computer science spans Theoretical computer science, theoretical disciplines (such as algorithms, theory of computation, and information theory) to Applied science, ...
, especially string searching algorithm
A string-searching algorithm, sometimes called string-matching algorithm, is an algorithm that searches a body of text for portions that match by pattern.
A basic example of string searching is when the pattern and the searched text are arrays of ...
s, machine learning
Machine learning (ML) is a field of study in artificial intelligence concerned with the development and study of Computational statistics, statistical algorithms that can learn from data and generalise to unseen data, and thus perform Task ( ...
, and database theory
Database theory encapsulates a broad range of topics related to the study and research of the theoretical realm of databases and database management systems.
Theoretical aspects of data management include, among other areas, the foundations of q ...
. String searching or matching algorithms, which find an occurrence of a sequence of letters inside a larger sequence of letters, were developed to search for specific sequences of nucleotides. The DNA sequence may be aligned with other DNA sequences to identify homologous sequences and locate the specific mutation
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, ...
s that make them distinct. These techniques, especially multiple sequence alignment
Multiple sequence alignment (MSA) is the process or the result of sequence alignment of three or more biological sequences, generally protein, DNA, or RNA. These alignments are used to infer evolutionary relationships via phylogenetic analysis an ...
, are used in studying phylogenetic
In biology, phylogenetics () is the study of the evolutionary history of life using observable characteristics of organisms (or genes), which is known as phylogenetic inference. It infers the relationship among organisms based on empirical dat ...
relationships and protein function. Data sets representing entire genomes' worth of DNA sequences, such as those produced by the Human Genome Project
The Human Genome Project (HGP) was an international scientific research project with the goal of determining the base pairs that make up human DNA, and of identifying, mapping and sequencing all of the genes of the human genome from both a ...
, are difficult to use without the annotations that identify the locations of genes and regulatory elements on each chromosome. Regions of DNA sequence that have the characteristic patterns associated with protein- or RNA-coding genes can be identified by gene finding algorithms, which allow researchers to predict the presence of particular gene product
A gene product is the biochemical material, either RNA or protein, resulting from the expression of a gene. A measurement of the amount of gene product is sometimes used to infer how active a gene is. Abnormal amounts of gene product can be corre ...
s and their possible functions in an organism even before they have been isolated experimentally.
DNA nanotechnology
 DNA nanotechnology uses the unique
DNA nanotechnology uses the unique molecular recognition
Supramolecular chemistry refers to the branch of chemistry concerning Chemical species, chemical systems composed of a integer, discrete number of molecules. The strength of the forces responsible for spatial organization of the system range from w ...
properties of DNA and other nucleic acids to create self-assembling branched DNA complexes with useful properties. DNA is thus used as a structural material rather than as a carrier of biological information. This has led to the creation of two-dimensional periodic lattices (both tile-based and using the ''DNA origami
DNA origami is the nanoscale folding of DNA to create arbitrary two- and three-dimensional shapes at the nanoscale. The specificity of the interactions between complementary base pairs make DNA a useful construction material, through design of i ...
'' method) and three-dimensional structures in the shapes of polyhedra
In geometry, a polyhedron (: polyhedra or polyhedrons; ) is a three-dimensional figure with flat polygonal faces, straight edges and sharp corners or vertices. The term "polyhedron" may refer either to a solid figure or to its boundary su ...
. Nanomechanical devices and algorithmic self-assembly have also been demonstrated, and these DNA structures have been used to template the arrangement of other molecules such as gold nanoparticles and streptavidin
Streptavidin is a 52 Atomic mass unit, kDa protein (tetramer) purified from the bacterium ''Streptomyces avidinii''. Streptavidin Homotetramer, homo-tetramers have an extraordinarily high affinity for biotin (also known as vitamin B7 or vitamin ...
proteins. DNA and other nucleic acids are the basis of aptamers, synthetic oligonucleotide ligands for specific target molecules used in a range of biotechnology and biomedical applications.
History and anthropology
Because DNA collects mutations over time, which are then inherited, it contains historical information, and, by comparing DNA sequences, geneticists can infer the evolutionary history of organisms, their phylogeny
A phylogenetic tree or phylogeny is a graphical representation which shows the evolutionary history between a set of species or Taxon, taxa during a specific time.Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, M ...
. This field of phylogenetics is a powerful tool in evolutionary biology
Evolutionary biology is the subfield of biology that studies the evolutionary processes such as natural selection, common descent, and speciation that produced the diversity of life on Earth. In the 1930s, the discipline of evolutionary biolo ...
. If DNA sequences within a species are compared, population geneticists
Population genetics is a subfield of genetics that deals with genetic differences within and among populations, and is a part of evolutionary biology. Studies in this branch of biology examine such phenomena as adaptation, speciation, and popula ...
can learn the history of particular populations. This can be used in studies ranging from ecological genetics
Ecological genetics is the study of genetics in natural populations. It combines ecology, evolution, and genetics to understand the processes behind adaptation. It is virtually synonymous with the field of molecular ecology.
This contrasts wit ...
to anthropology
Anthropology is the scientific study of humanity, concerned with human behavior, human biology, cultures, society, societies, and linguistics, in both the present and past, including archaic humans. Social anthropology studies patterns of behav ...
.
Information storage
DNA as a storage device for information has enormous potential since it has much higher storage density compared to electronic devices. However, high costs, slow read and write times (memory latency
''Memory latency'' is the time (the latency) between initiating a request for a byte or word in memory until it is retrieved by a processor. If the data are not in the processor's cache, it takes longer to obtain them, as the processor will ha ...
), and insufficient reliability
Reliability, reliable, or unreliable may refer to:
Science, technology, and mathematics Computing
* Data reliability (disambiguation), a property of some disk arrays in computer storage
* Reliability (computer networking), a category used to des ...
has prevented its practical use.
History
 DNA was first isolated by the Swiss physician
DNA was first isolated by the Swiss physician Friedrich Miescher
Johannes Friedrich Miescher (13 August 1844 – 26 August 1895) was a Swiss physician and biologist. He was the first scientist to isolate nucleic acid in 1869. Miescher also identified protamine and made several other discoveries.
Miescher had ...
who, in 1869, discovered a microscopic substance in the pus
Pus is an exudate, typically white-yellow, yellow, or yellow-brown, formed at the site of inflammation during infections, regardless of cause. An accumulation of pus in an enclosed tissue space is known as an abscess, whereas a visible collect ...
of discarded surgical bandages. As it resided in the nuclei of cells, he called it "nuclein". In 1878, Albrecht Kossel
Ludwig Karl Martin Leonhard Albrecht Kossel (; 16 September 1853 – 5 July 1927) was a biochemist and pioneer in the study of genetics. He was awarded the Nobel Prize for Physiology or Medicine in 1910 for his work in determining the chemical ...
isolated the non-protein component of "nuclein", nucleic acid, and later isolated its five primary nucleobase
Nucleotide bases (also nucleobases, nitrogenous bases) are nitrogen-containing biological compounds that form nucleosides, which, in turn, are components of nucleotides, with all of these monomers constituting the basic building blocks of nuc ...
s.Phoebus Levene
Phoebus Aaron Theodore Levene (25 February 1869 – 6 September 1940) was a Russian-born American biochemist who studied the structure and function of nucleic acids. He characterized the different forms of nucleic acid, DNA from RNA, and foun ...
identified the base, sugar, and phosphate nucleotide unit of RNA (then named "yeast nucleic acid"). In 1929, Levene identified deoxyribose sugar in "thymus nucleic acid" (DNA). Levene suggested that DNA consisted of a string of four nucleotide units linked together through the phosphate groups ("tetranucleotide hypothesis"). Levene thought the chain was short and the bases repeated in a fixed order. In 1927, Nikolai Koltsov
Nikolai Konstantinovich Koltsov (; 14 July 1872 – 2 December 1940) was a Russian biologist and a pioneer of modern genetics. Among his students were Nikolay Timofeeff-Ressovsky, Vladimir Pavlovich Efroimson, Alexander Sergeevich Serebrovsky, A ...
proposed that inherited traits would be inherited via a "giant hereditary molecule" made up of "two mirror strands that would replicate in a semi-conservative fashion using each strand as a template".Frederick Griffith
Frederick Griffith (1877–1941) was a British bacteriologist whose focus was the epidemiology and pathology of bacterial pneumonia. In January 1928 he reported what is now known as Griffith's experiment, the first widely accepted demonstrat ...
in his experiment
An experiment is a procedure carried out to support or refute a hypothesis, or determine the efficacy or likelihood of something previously untried. Experiments provide insight into cause-and-effect by demonstrating what outcome occurs whe ...
discovered that traits
Trait may refer to:
* Phenotypic trait in biology, which involve genes and characteristics of organisms
* Genotypic trait, sometimes but not always presenting as a phenotypic trait
* Personality, traits that predict an individual's behavior.
** ...
of the "smooth" form of ''Pneumococcus'' could be transferred to the "rough" form of the same bacteria by mixing killed "smooth" bacteria with the live "rough" form. This system provided the first clear suggestion that DNA carries genetic information.
In 1933, while studying virgin sea urchin
Sea urchins or urchins () are echinoderms in the class (biology), class Echinoidea. About 950 species live on the seabed, inhabiting all oceans and depth zones from the intertidal zone to deep seas of . They typically have a globular body cove ...
eggs, Jean Brachet
Jean Louis Auguste Brachet (19 March 1909 – 10 August 1988) was a Belgian biochemist who made a key contribution in understanding the role of RNA.
Life
Brachet was born in Etterbeek near Brussels in Belgium, the son of Albert Brachet, a ...
suggested that DNA is found in the cell nucleus
The cell nucleus (; : nuclei) is a membrane-bound organelle found in eukaryote, eukaryotic cell (biology), cells. Eukaryotic cells usually have a single nucleus, but a few cell types, such as mammalian red blood cells, have #Anucleated_cells, ...
and that RNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA and deoxyrib ...
is present exclusively in the cytoplasm
The cytoplasm describes all the material within a eukaryotic or prokaryotic cell, enclosed by the cell membrane, including the organelles and excluding the nucleus in eukaryotic cells. The material inside the nucleus of a eukaryotic cell a ...
. At the time, "yeast nucleic acid" (RNA) was thought to occur only in plants, while "thymus nucleic acid" (DNA) only in animals. The latter was thought to be a tetramer, with the function of buffering cellular pH.
In 1937, William Astbury
William Thomas Astbury FRS (25 February 1898 – 4 June 1961) was an English physicist and molecular biologist who made pioneering X-ray diffraction studies of biological molecules. His work on keratin provided the foundation for Linus Pauli ...
produced the first X-ray diffraction patterns that showed that DNA had a regular structure.
In 1943, Oswald Avery
Oswald Theodore Avery Jr. (October 21, 1877 – February 20, 1955) was a Canadian-American physician and medical researcher. The major part of his career was spent at the Rockefeller Hospital in New York City. Avery was one of the first molecu ...
, along with co-workers Colin MacLeod and Maclyn McCarty
Maclyn McCarty (June 9, 1911 – January 2, 2005) was an American geneticist, a research scientist described in 2005 as "the last surviving member of a Manhattan scientific team that overturned medical dogma in the 1940s and became the first to ...
, identified DNA as the transforming principle, supporting Griffith's suggestion (Avery–MacLeod–McCarty experiment
The Avery–MacLeod–McCarty experiment was an experimental demonstration by Oswald Avery, Colin MacLeod, and Maclyn McCarty that, in 1944, reported that DNA is the substance that causes bacterial transformation, in an era when it had been wi ...
). Erwin Chargaff
Erwin Chargaff (11 August 1905 – 20 June 2002) was an Austro-Hungarian-born American biochemist, writer, and professor of biochemistry at Columbia University medical school. A Bucovinian Jew who immigrated to the United States during the Nazi ...
developed and published observations now known as Chargaff's rules
Chargaff's rules (given by Erwin Chargaff) state that in the DNA of any species and any organism, the amount of guanine should be equal to the amount of cytosine and the amount of adenine should be equal to the amount of thymine. Further, a 1:1 st ...
, stating that in DNA from any species of any organism, the amount of guanine
Guanine () (symbol G or Gua) is one of the four main nucleotide bases found in the nucleic acids DNA and RNA, the others being adenine, cytosine, and thymine ( uracil in RNA). In DNA, guanine is paired with cytosine. The guanine nucleoside ...
should be equal to cytosine
Cytosine () (symbol C or Cyt) is one of the four nucleotide bases found in DNA and RNA, along with adenine, guanine, and thymine ( uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attac ...
and the amount of adenine
Adenine (, ) (nucleoside#List of nucleosides and corresponding nucleobases, symbol A or Ade) is a purine nucleotide base that is found in DNA, RNA, and Adenosine triphosphate, ATP. Usually a white crystalline subtance. The shape of adenine is ...
should be equal to thymine
Thymine () (symbol T or Thy) is one of the four nucleotide bases in the nucleic acid of DNA that are represented by the letters G–C–A–T. The others are adenine, guanine, and cytosine. Thymine is also known as 5-methyluracil, a pyrimidine ...
. Late in 1951,
Late in 1951, Francis Crick
Francis Harry Compton Crick (8 June 1916 – 28 July 2004) was an English molecular biologist, biophysicist, and neuroscientist. He, James Watson, Rosalind Franklin, and Maurice Wilkins played crucial roles in deciphering the Nucleic acid doub ...
started working with James Watson
James Dewey Watson (born April 6, 1928) is an American molecular biology, molecular biologist, geneticist, and zoologist. In 1953, he co-authored with Francis Crick the academic paper in ''Nature (journal), Nature'' proposing the Nucleic acid ...
at the Cavendish Laboratory
The Cavendish Laboratory is the Department of Physics at the University of Cambridge, and is part of the School of Physical Sciences. The laboratory was opened in 1874 on the New Museums Site as a laboratory for experimental physics and is named ...
within the University of Cambridge
The University of Cambridge is a Public university, public collegiate university, collegiate research university in Cambridge, England. Founded in 1209, the University of Cambridge is the List of oldest universities in continuous operation, wo ...
. DNA's role in heredity
Heredity, also called inheritance or biological inheritance, is the passing on of traits from parents to their offspring; either through asexual reproduction or sexual reproduction, the offspring cells or organisms acquire the genetic infor ...
was confirmed in 1952 when Alfred Hershey
Alfred Day Hershey (December 4, 1908 – May 22, 1997) was an American Nobel Prize–winning bacteriologist and geneticist.
Early years
Hershey was born in Owosso, Michigan to Robert Day and Alma Wilbur Hershey. He earned a B.S. in chemistr ...
and Martha Chase in the Hershey–Chase experiment
The Hershey–Chase experiments were a series of experiments conducted in 1952 by Alfred Hershey and Martha Chase that helped to confirm that DNA is genetic material.
While DNA had been known to biologists since 1869, many scientists still as ...
showed that DNA is the genetic material
Nucleic acids are large biomolecules that are crucial in all cells and viruses. They are composed of nucleotides, which are the monomer components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main classes of nucleic aci ...
of the enterobacteria phage T2.
In May 1952, Raymond Gosling
Raymond George Gosling (15 July 1926 – 18 May 2015) was a British scientist. While a PhD student at King's College, London he worked under the supervision of Maurice Wilkins and Rosalind Franklin. The crystallographic experiments of Frankl ...
, a graduate student working under the supervision of Rosalind Franklin
Rosalind Elsie Franklin (25 July 192016 April 1958) was a British chemist and X-ray crystallographer. Her work was central to the understanding of the molecular structures of DNA (deoxyribonucleic acid), RNA (ribonucleic acid), viruses, coal ...
, took an X-ray diffraction
X-ray diffraction is a generic term for phenomena associated with changes in the direction of X-ray beams due to interactions with the electrons around atoms. It occurs due to elastic scattering, when there is no change in the energy of the waves. ...
image, labeled as "Photo 51 '
''Photo 51'' is an X-ray diffraction, X-ray based fiber diffraction image of a paracrystalline gel composed of DNA fiber taken by Raymond Gosling, a postgraduate student working under the supervision of Maurice Wilkins and Rosalind Franklin ...
", at high hydration levels of DNA. This photo was given to Watson and Crick by Maurice Wilkins
Maurice Hugh Frederick Wilkins (15 December 1916 – 5 October 2004) was a New Zealand-born British biophysicist and Nobel laureate whose research spanned multiple areas of physics and biophysics, contributing to the scientific understanding ...
and was critical to their obtaining the correct structure of DNA. Franklin told Crick and Watson that the backbones had to be on the outside. Before then, Linus Pauling, and Watson and Crick, had erroneous models with the chains inside and the bases pointing outwards. Franklin's identification of the space group
In mathematics, physics and chemistry, a space group is the symmetry group of a repeating pattern in space, usually in three dimensions. The elements of a space group (its symmetry operations) are the rigid transformations of the pattern that ...
for DNA crystals revealed to Crick that the two DNA strands were antiparallel.Linus Pauling
Linus Carl Pauling ( ; February 28, 1901August 19, 1994) was an American chemist and peace activist. He published more than 1,200 papers and books, of which about 850 dealt with scientific topics. ''New Scientist'' called him one of the 20 gre ...
and Robert Corey
Robert Brainard Corey (August 19, 1897 – April 23, 1971) was an American biochemist, mostly known for his role in discovery of the α-helix and the β-sheet with Linus Pauling. Also working with Pauling was Herman Branson. Their discoveries ...
proposed a model for nucleic acids containing three intertwined chains, with the phosphates near the axis, and the bases on the outside.DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
. On 28 February 1953 Crick interrupted patrons' lunchtime at The Eagle pub
A pub (short for public house) is in several countries a drinking establishment licensed to serve alcoholic drinks for consumption on the premises. The term first appeared in England in the late 17th century, to differentiate private ho ...
in Cambridge, England to announce that he and Watson had "discovered the secret of life".
 The 25 April 1953 issue of the journal ''
The 25 April 1953 issue of the journal ''Nature
Nature is an inherent character or constitution, particularly of the Ecosphere (planetary), ecosphere or the universe as a whole. In this general sense nature refers to the Scientific law, laws, elements and phenomenon, phenomena of the physic ...
'' published a series of five articles giving the Watson and Crick double-helix structure DNA and evidence supporting it.Nobel Prize in Physiology or Medicine
The Nobel Prize in Physiology or Medicine () is awarded yearly by the Nobel Assembly at the Karolinska Institute for outstanding discoveries in physiology or medicine. The Nobel Prize is not a single prize, but five separate prizes that, acco ...
. Nobel Prizes are awarded only to living recipients. A debate continues about who should receive credit for the discovery.
In an influential presentation in 1957, Crick laid out the central dogma of molecular biology
The central dogma of molecular biology deals with the flow of genetic information within a biological system. It is often stated as "DNA makes RNA, and RNA makes protein", although this is not its original meaning. It was first stated by Francis Cr ...
, which foretold the relationship between DNA, RNA, and proteins, and articulated the "adaptor hypothesis". Final confirmation of the replication mechanism that was implied by the double-helical structure followed in 1958 through the Meselson–Stahl experiment. Further work by Crick and co-workers showed that the genetic code was based on non-overlapping triplets of bases, called codons
Genetic code is a set of rules used by living cells to translate information encoded within genetic material ( DNA or RNA sequences of nucleotide triplets or codons) into proteins. Translation is accomplished by the ribosome, which links pro ...
, allowing Har Gobind Khorana
Har Gobind Khorana (9 January 1922 – 9 November 2011) was an Indian-American biochemist. While on the faculty of the University of Wisconsin–Madison, he shared the 1968 Nobel Prize for Physiology or Medicine with Marshall W. Nirenberg and ...
, Robert W. Holley, and Marshall Warren Nirenberg to decipher the genetic code. These findings represent the birth of molecular biology
Molecular biology is a branch of biology that seeks to understand the molecule, molecular basis of biological activity in and between Cell (biology), cells, including biomolecule, biomolecular synthesis, modification, mechanisms, and interactio ...
.
In 1986, DNA analysis was first used in a criminal investigation when police in the UK requested Alec Jeffreys
Sir Alec John Jeffreys, (born 9 January 1950) is a British geneticist known for developing techniques for genetic fingerprinting and DNA profiling which are now used worldwide in forensic science to assist police detective work and to resolve ...
of the University of Leicester to prove or disprove the involvement in a particular case of a suspect who claimed innocence in the matter. Although the suspect had already confessed to committing a recent rape-murder, he was denying any involvement in a similar crime committed three years earlier. Yet the details of the two cases were so alike that the police concluded both crimes had been committed by the same person. However, all charges against the suspect were dropped when Jeffreys' DNA testing exonerated the suspect — from both the earlier murder and the one to which he'd confessed. Further such DNA profiling led to positive identification of another suspect who, in 1988, was found guilty of both rape-murders.
See also
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
*
References
Further reading
*
*
*
*
* First published in October 1974 by MacMillan, with foreword by Francis Crick; the definitive DNA textbook, revised in 1994 with a nine-page postscript.
*
*
*
*
*
*
*
*
*
External links
DNA binding site prediction on protein
DNA the Double Helix Game
From the official Nobel Prize web site
Dolan DNA Learning Center
''Nature
Nature is an inherent character or constitution, particularly of the Ecosphere (planetary), ecosphere or the universe as a whole. In this general sense nature refers to the Scientific law, laws, elements and phenomenon, phenomena of the physic ...
''
*
*
ENCODE threads explorer
ENCODE home page at Nature
Nature is an inherent character or constitution, particularly of the Ecosphere (planetary), ecosphere or the universe as a whole. In this general sense nature refers to the Scientific law, laws, elements and phenomenon, phenomena of the physic ...
Double Helix 1953–2003
National Centre for Biotechnology Education
Genetic Education Modules for Teachers
– ''DNA from the Beginning'' Study Guide
*
"Clue to chemistry of heredity found"
''The New York Times'', June 1953. First American newspaper coverage of the discovery of the DNA structure
DNA from the Beginning
Another DNA Learning Center site on DNA, genes, and heredity from Mendel to the human genome project.
at Mandeville Special Collections Library, University of California, San Diego
Seven-page, handwritten letter that Crick sent to his 12-year-old son Michael in 1953 describing the structure of DNA.
Se
Crick's medal goes under the hammer
Nature, 5 April 2013.
{{Authority control
DNA,
Biotechnology
Helices
Nucleic acids
 The DNA double helix is stabilized primarily by two forces:
The DNA double helix is stabilized primarily by two forces:  Twin helical strands form the DNA backbone. Another double helix may be found tracing the spaces, or grooves, between the strands. These voids are adjacent to the base pairs and may provide a
Twin helical strands form the DNA backbone. Another double helix may be found tracing the spaces, or grooves, between the strands. These voids are adjacent to the base pairs and may provide a  In humans, the total female
In humans, the total female  DNA exists in many possible conformations that include
DNA exists in many possible conformations that include  At the ends of the linear chromosomes are specialized regions of DNA called
At the ends of the linear chromosomes are specialized regions of DNA called  DNA can be damaged by many sorts of
DNA can be damaged by many sorts of  Some noncoding DNA sequences play structural roles in chromosomes.
Some noncoding DNA sequences play structural roles in chromosomes.  Structural proteins that bind DNA are well-understood examples of non-specific DNA-protein interactions. Within chromosomes, DNA is held in complexes with structural proteins. These proteins organize the DNA into a compact structure called
Structural proteins that bind DNA are well-understood examples of non-specific DNA-protein interactions. Within chromosomes, DNA is held in complexes with structural proteins. These proteins organize the DNA into a compact structure called  In contrast, other proteins have evolved to bind to particular DNA sequences. The most intensively studied of these are the various
In contrast, other proteins have evolved to bind to particular DNA sequences. The most intensively studied of these are the various 
 A DNA helix usually does not interact with other segments of DNA, and in human cells, the different chromosomes even occupy separate areas in the nucleus called "
A DNA helix usually does not interact with other segments of DNA, and in human cells, the different chromosomes even occupy separate areas in the nucleus called " DNA nanotechnology uses the unique
DNA nanotechnology uses the unique  DNA was first isolated by the Swiss physician
DNA was first isolated by the Swiss physician  Late in 1951,
Late in 1951,  The 25 April 1953 issue of the journal ''
The 25 April 1953 issue of the journal ''