|
Hoogsteen Base Pair
A Hoogsteen base pair is a variation of base-pairing in nucleic acids such as the A•T pair. In this manner, two nucleobases, one on each strand, can be held together by hydrogen bonds in the major groove. A Hoogsteen base pair applies the N7 position of the purine base (as a hydrogen bond acceptor) and C6 amino group (as a donor), which bind the Watson–Crick (N3–C4) face of the pyrimidine base. History Ten years after James Watson and Francis Crick published their model of the DNA double helix, Karst Hoogsteen reported a crystal structure of a complex in which analogues of A and T formed a base pair that had a different geometry from that described by Watson and Crick. Similarly, an alternative base-pairing geometry can occur for G•C pairs. Hoogsteen pointed out that if the alternative hydrogen-bonding patterns were present in DNA, then the double helix would have to assume a quite different shape. Hoogsteen base pairs are observed in alternative structures such ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hoogsteen Watson Crick Pairing-en
A Hoogsteen base pair is a variation of base-pairing in nucleic acids such as the A•T pair. In this manner, two nucleobases, one on each strand, can be held together by hydrogen bonds in the major groove. A Hoogsteen base pair applies the N7 position of the purine base (as a hydrogen bond acceptor) and C6 amino group (as a donor), which bind the Watson–Crick (N3–C4) face of the pyrimidine base. History Ten years after James Watson and Francis Crick published their model of the DNA double helix, Karst Hoogsteen reported a crystal structure of a complex in which analogues of A and T formed a base pair that had a different geometry from that described by Watson and Crick. Similarly, an alternative base-pairing geometry can occur for G•C pairs. Hoogsteen pointed out that if the alternative hydrogen-bonding patterns were present in DNA, then the double helix would have to assume a quite different shape. Hoogsteen base pairs are observed in alternative structures such as the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
G-quadruplex
In molecular biology, G-quadruplex secondary structures (G4) are formed in nucleic acids by sequences that are rich in guanine. They are helical in shape and contain guanine tetrads that can form from one, two or four strands. The unimolecular forms often occur naturally near the ends of the chromosomes, better known as the telomeric regions, and in transcriptional regulatory regions of multiple genes, both in microbes and across vertebrates including oncogenes in humans. Four guanine bases can associate through Hoogsteen hydrogen bonding to form a square planar structure called a guanine tetrad (G-tetrad or G-quartet), and two or more guanine tetrads (from G-tracts, continuous runs of guanine) can stack on top of each other to form a G-quadruplex. The placement and bonding to form G-quadruplexes is not random and serve very unusual functional purposes. The quadruplex structure is further stabilized by the presence of a cation, especially potassium, which sits in a central c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
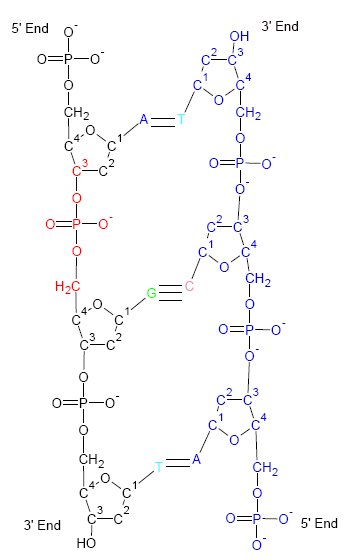
Nucleic Acid Tertiary Structure
Nucleic acid tertiary structure is the three-dimensional shape of a nucleic acid polymer. RNA and DNA molecules are capable of diverse functions ranging from molecular recognition to catalysis. Such functions require a precise three-dimensional structure. While such structures are diverse and seemingly complex, they are composed of recurring, easily recognizable tertiary structural motifs that serve as molecular building blocks. Some of the most common motifs for RNA and DNA tertiary structure are described below, but this information is based on a limited number of solved structures. Many more tertiary structural motifs will be revealed as new RNA and DNA molecules are structurally characterized. Helical structures Double helix The double helix is the dominant tertiary structure for biological DNA, and is also a possible structure for RNA. Three DNA conformations are believed to be found in nature, A-DNA, B-DNA, and Z-DNA. The "B" form described by James D. Wats ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Guanine Tetrad
In molecular biology, a guanine tetrad (also known as a G-tetrad or G-quartet) is a structure composed of four guanine bases in a square planar array. They most prominently contribute to the structure of G-quadruplexes, where their hydrogen bonding stabilizes the structure. Usually, there are at least two guanine tetrads in a G-quadruplex, and they often feature Hoogsteen-style hydrogen bonding. Guanine tetrads are formed by sequences rich in guanine, such as GGGGC. They may also play a role in the dimerization of non-endogenous RNAs to facilitate the replication of some viruses. Guanine tetrads dimerize through their 5' ends since it is more energetically favorable. They can be stabilized by central cations, such as lithium, sodium, potassium, rubidium, or cesium. However, they still form a variety of different structures. Guanine tetrads are not always stable, but the sugar-phosphate backbone of DNA can assist in stability of the guanine tetrads themselves. Guanine tetra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Wobble Base Pair
A wobble base pair is a pairing between two nucleotides in RNA molecules that does not follow Watson-Crick base pair rules. The four main wobble base pairs are guanine-uracil (G-U), hypoxanthine-uracil (I-U), hypoxanthine-adenine (I-A), and hypoxanthine-cytosine (I-C). In order to maintain consistency of nucleic acid nomenclature, "I" is used for hypoxanthine because hypoxanthine is the nucleobase of inosine; nomenclature otherwise follows the names of nucleobases and their corresponding nucleosides (e.g., "G" for both guanine and guanosine – as well as for deoxyguanosine). The thermodynamic stability of a wobble base pair is comparable to that of a Watson-Crick base pair. Wobble base pairs are fundamental in RNA secondary structure and are critical for the proper translation of the genetic code. Brief history In the genetic code, there are 43 = 64 possible codons (3 nucleotide sequences). For translation, each of these codons requires a tRNA molecule with an anticodon with whic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transfer RNA
Transfer RNA (abbreviated tRNA and formerly referred to as sRNA, for soluble RNA) is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length (in eukaryotes), that serves as the physical link between the mRNA and the amino acid sequence of proteins. tRNAs genes from Bacteria are typically shorter (mean = 77.6 bp) than tRNAs from Archaea (mean = 83.1 bp) and eukaryotes (mean = 84.7 bp). The mature tRNA follows an opposite pattern with tRNAs from Bacteria being usually longer (median = 77.6 nt) than tRNAs from Archaea (median = 76.8 nt), with eukaryotes exhibiting the shortest mature tRNAs (median = 74.5 nt). Transfer RNA (tRNA) does this by carrying an amino acid to the protein synthesizing machinery of a cell called the ribosome. Complementation of a 3-nucleotide codon in a messenger RNA (mRNA) by a 3-nucleotide anticodon of the tRNA results in protein synthesis based on the mRNA code. As such, tRNAs are a necessary component of translation, the biological sy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Triple-stranded DNA
Triple-stranded DNA (also known as H-DNA or Triplex-DNA) is a DNA structure in which three oligonucleotides wind around each other and form a triple helix. In triple-stranded DNA, the third strand binds to a B-form DNA (via Watson–Crick base-pairing) double helix by forming Hoogsteen base pairs or reversed Hoogsteen hydrogen bonds. Structure Examples of triple-stranded DNA have been found in natural sources with the required structural elements, for example in Satellite DNA. Hoogsteen base pairing A thymine (T) nucleobase can bind to a Watson–Crick base-pairing of T-A by forming a Hoogsteen hydrogen bond. The thymine hydrogen bonds with the adenosine (A) of the original double-stranded DNA to create a T-A*T base-triplet. Intermolecular and intramolecular interactions There are two classes of triplex DNA: intermolecular and intramolecular formations. An intermolecular triplex refers to triplex formation between a duplex and a different (third) strand of DNA. Th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hoogsteen
A Hoogsteen base pair is a variation of base-pairing in nucleic acids such as the A•T pair. In this manner, two nucleobases, one on each strand, can be held together by hydrogen bonds in the major groove. A Hoogsteen base pair applies the N7 position of the purine base (as a hydrogen bond acceptor) and C6 amino group (as a donor), which bind the Watson–Crick (N3–C4) face of the pyrimidine base. History Ten years after James Watson and Francis Crick published their model of the DNA double helix, Karst Hoogsteen reported a crystal structure of a complex in which analogues of A and T formed a base pair that had a different geometry from that described by Watson and Crick. Similarly, an alternative base-pairing geometry can occur for G•C pairs. Hoogsteen pointed out that if the alternative hydrogen-bonding patterns were present in DNA, then the double helix would have to assume a quite different shape. Hoogsteen base pairs are observed in alternative structures such ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tamar Schlick
Tamar Schlick is an American applied mathematician who works as a professor of chemistry, mathematics, and computer science at New York University. Her research involves developing and applying tools for modeling and simulating biomolecules.. Education and career Schlick did her undergraduate studies at Wayne State University, graduating in 1982 with a B.S. in mathematics. She continued her graduate studies at the Courant Institute of Mathematical Sciences at New York University, completing a Ph.D. in applied mathematics in 1987 under the supervision of Charles S. Peskin. After postdoctoral studies at NYU and the Weizmann Institute of Science, she returned as a faculty member to NYU in 1989. Recognition She is a fellow of the American Association for the Advancement of Science (2004), American Physical Society (2005), Biophysical Society (2012), and Society for Industrial and Applied Mathematics Society for Industrial and Applied Mathematics (SIAM) is a professional society d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Karst Hoogsteen
Karst Hoogsteen (October 1, 1923 – August 10, 2015) was a Dutch-born American biochemist famous for noting a new base pairing form in DNA, now called Hoogsteen base pairs.. These base pairing intercede in the Watson-Crick base pairing A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DN ..., forging a base pair 'triplex'. The Base Pairs use the N7 nitrogen atom as the accepter rather than the N1 as observed in Watson-Crick base pairing. This leads to a twisted, non-linear arrangement. References 1923 births 2015 deaths American biochemists Dutch biochemists Dutch emigrants to the United States University of Groningen alumni Scientists from Groningen (city) {{US-biochemist-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleic Acid
Nucleic acids are biopolymers, macromolecules, essential to all known forms of life. They are composed of nucleotides, which are the monomers made of three components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main classes of nucleic acids are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). If the sugar is ribose, the polymer is RNA; if the sugar is the ribose derivative deoxyribose, the polymer is DNA. Nucleic acids are naturally occurring chemical compounds that serve as the primary information-carrying molecules in cells and make up the genetic material. Nucleic acids are found in abundance in all living things, where they create, encode, and then store information of every living cell of every life-form on Earth. In turn, they function to transmit and express that information inside and outside the cell nucleus to the interior operations of the cell and ultimately to the next generation of each living organism. The encoded information is ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Francis Crick
Francis Harry Compton Crick (8 June 1916 – 28 July 2004) was an English molecular biologist, biophysicist, and neuroscientist. He, James Watson, Rosalind Franklin, and Maurice Wilkins played crucial roles in deciphering the helical structure of the DNA molecule. Crick and Watson's paper in ''Nature'' in 1953 laid the groundwork for understanding DNA structure and functions. Together with Maurice Wilkins, they were jointly awarded the 1962 Nobel Prize in Physiology or Medicine "for their discoveries concerning the molecular structure of nucleic acids and its significance for information transfer in living material". Crick was an important theoretical molecular biologist and played a crucial role in research related to revealing the helical structure of DNA. He is widely known for the use of the term " central dogma" to summarise the idea that once information is transferred from nucleic acids (DNA or RNA) to proteins, it cannot flow back to nucleic acids. In other words ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |

.png)