|
Multiple Sequence Alignment
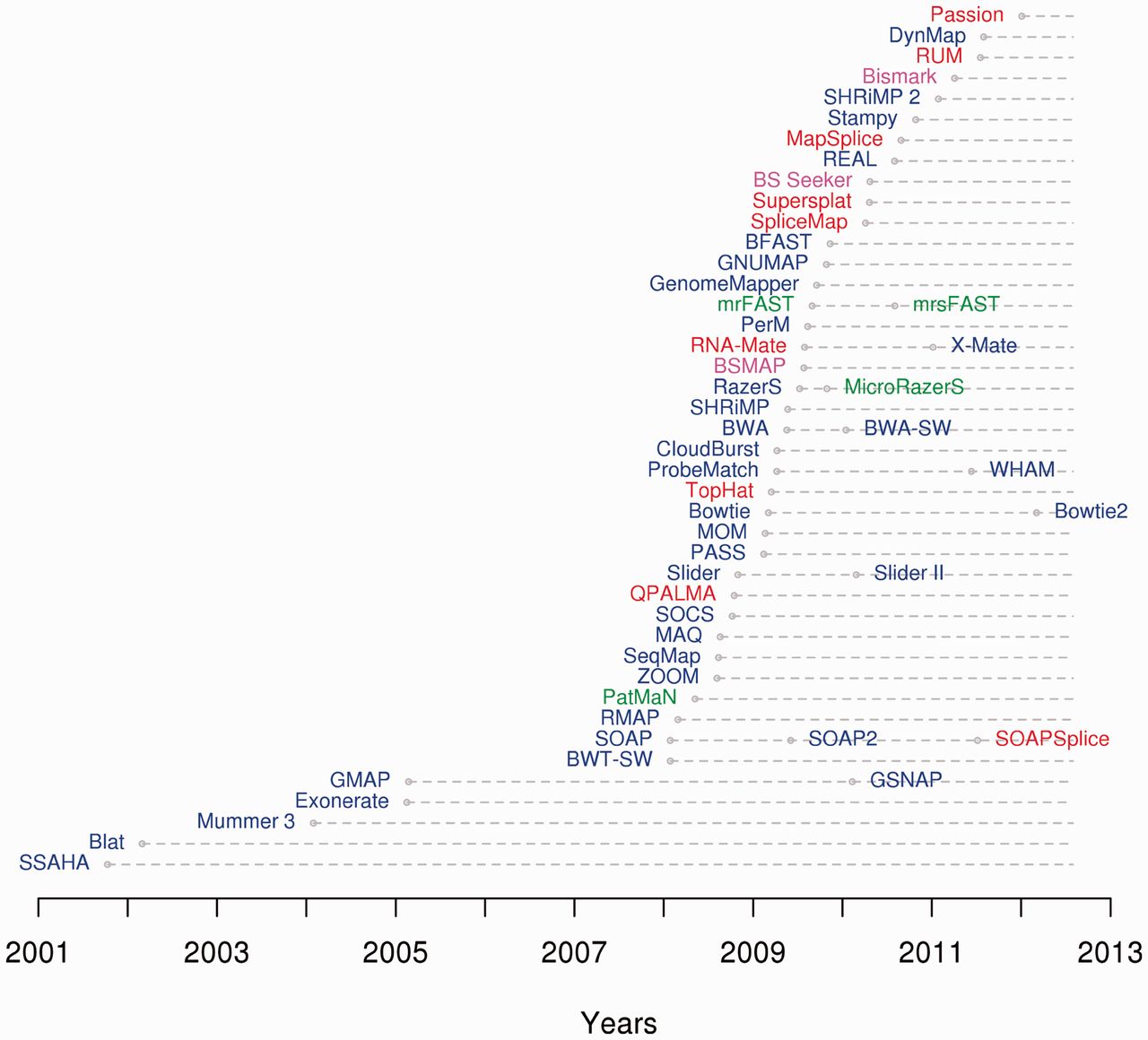
Multiple sequence alignment (MSA) may refer to the process or the result of sequence alignment of three or more biological sequences, generally protein, DNA, or RNA. In many cases, the input set of query sequences are assumed to have an evolutionary relationship by which they share a linkage and are descended from a common ancestor. From the resulting MSA, sequence homology can be inferred and phylogenetic analysis can be conducted to assess the sequences' shared evolutionary origins. Visual depictions of the alignment as in the image at right illustrate mutation events such as point mutations (single amino acid or nucleotide changes) that appear as differing characters in a single alignment column, and insertion or deletion mutations (indels or gaps) that appear as hyphens in one or more of the sequences in the alignment. Multiple sequence alignment is often used to assess sequence conservation of protein domains, tertiary and secondary structures, and even individual amino acid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RPLP0 90 ClustalW Aln
Legazpi Airport (Filipino: ''Paliparan ng Legazpi'', Bikol: '' Palayogan nin Legazpi'', ) was a major airport in the Bicol Region, served the vicinity of Legazpi, the capital city of Albay in the Philippines. Prior to its closure, the airport was designated as a Class 1 principal domestic Airport by the Civil Aviation Authority of the Philippines, a body of the Department of Transportation that is responsible for the operations of not only this airport but also of all other public airports in the Philippines except the major international airports. The airport was replaced by the new and larger Bicol International Airport in 2021. History The site of the former Legazpi Airport used to be the Sanborn Field. This former US airfield opened in the early 1900s. During World War II, the airport was occupied by Japanese forces from 1941 until the airport was liberated by the United States Army in 1945. Commercial operations started the following year. The jet age came to Legazp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heuristic
A heuristic (; ), or heuristic technique, is any approach to problem solving or self-discovery that employs a practical method that is not guaranteed to be optimal, perfect, or rational, but is nevertheless sufficient for reaching an immediate, short-term goal or approximation. Where finding an optimal solution is impossible or impractical, heuristic methods can be used to speed up the process of finding a satisfactory solution. Heuristics can be mental shortcuts that ease the cognitive load of making a decision. Examples that employ heuristics include using trial and error, a rule of thumb or an educated guess. Heuristics are the strategies derived from previous experiences with similar problems. These strategies depend on using readily accessible, though loosely applicable, information to control problem solving in human beings, machines and abstract issues. When an individual applies a heuristic in practice, it generally performs as expected. However it can alternati ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FASTA
FASTA is a DNA and protein sequence alignment software package first described by David J. Lipman and William R. Pearson in 1985. Its legacy is the FASTA format which is now ubiquitous in bioinformatics. History The original FASTA program was designed for protein sequence similarity searching. Because of the exponentially expanding genetic information and the limited speed and memory of computers in the 1980s heuristic methods were introduced aligning a query sequence to entire data-bases. FASTA, published in 1987, added the ability to do DNA:DNA searches, translated protein:DNA searches, and also provided a more sophisticated shuffling program for evaluating statistical significance. There are several programs in this package that allow the alignment of protein sequences and DNA sequences. Nowadays, increased computer performance makes it possible to perform searches for local alignment detection in a database using the Smith–Waterman algorithm. FASTA is pronounced "f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
UPGMA
UPGMA (unweighted pair group method with arithmetic mean) is a simple agglomerative (bottom-up) hierarchical clustering method. The method is generally attributed to Sokal and Michener. The UPGMA method is similar to its ''weighted'' variant, the WPGMA method. Note that the unweighted term indicates that all distances contribute equally to each average that is computed and does not refer to the math by which it is achieved. Thus the simple averaging in WPGMA produces a weighted result and the proportional averaging in UPGMA produces an unweighted result ('' see the working example''). Algorithm The UPGMA algorithm constructs a rooted tree (dendrogram) that reflects the structure present in a pairwise similarity matrix (or a dissimilarity matrix). At each step, the nearest two clusters are combined into a higher-level cluster. The distance between any two clusters \mathcal and \mathcal, each of size (''i.e.'', cardinality) and , is taken to be the average of all distances d(x, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Neighbor-joining
In bioinformatics, neighbor joining is a bottom-up (agglomerative) clustering method for the creation of phylogenetic trees, created by Naruya Saitou and Masatoshi Nei in 1987. Usually based on DNA or protein sequence data, the algorithm requires knowledge of the distance between each pair of taxa (e.g., species or sequences) to create the phylogenetic tree. The algorithm Neighbor joining takes a distance matrix, which specifies the distance between each pair of taxa, as input. The algorithm starts with a completely unresolved tree, whose topology corresponds to that of a star network, and iterates over the following steps, until the tree is completely resolved, and all branch lengths are known: # Based on the current distance matrix, calculate a matrix Q (defined below). # Find the pair of distinct taxa i and j (i.e. with i \neq j) for which Q(i,j) is smallest. Make a new node that joins the taxa i and j, and connect the new node to the central node. For example, in part (B ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cluster Analysis
Cluster analysis or clustering is the task of grouping a set of objects in such a way that objects in the same group (called a cluster) are more similar (in some sense) to each other than to those in other groups (clusters). It is a main task of exploratory data analysis, and a common technique for statistical data analysis, used in many fields, including pattern recognition, image analysis, information retrieval, bioinformatics, data compression, computer graphics and machine learning. Cluster analysis itself is not one specific algorithm, but the general task to be solved. It can be achieved by various algorithms that differ significantly in their understanding of what constitutes a cluster and how to efficiently find them. Popular notions of clusters include groups with small distances between cluster members, dense areas of the data space, intervals or particular statistical distributions. Clustering can therefore be formulated as a multi-objective optimization probl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylogenetic Tree
A phylogenetic tree (also phylogeny or evolutionary tree Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA.) is a branching diagram or a tree showing the evolutionary relationships among various biological species or other entities based upon similarities and differences in their physical or genetic characteristics. All life on Earth is part of a single phylogenetic tree, indicating common ancestry. In a ''rooted'' phylogenetic tree, each node with descendants represents the inferred most recent common ancestor of those descendants, and the edge lengths in some trees may be interpreted as time estimates. Each node is called a taxonomic unit. Internal nodes are generally called hypothetical taxonomic units, as they cannot be directly observed. Trees are useful in fields of biology such as bioinformatics, systematics, and phylogenetics. ''Unrooted'' trees illustrate only the relatedness of the leaf nodes and do not require the ancestral root ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NP Complete
In computational complexity theory, a problem is NP-complete when: # it is a problem for which the correctness of each solution can be verified quickly (namely, in polynomial time) and a brute-force search algorithm can find a solution by trying all possible solutions. # the problem can be used to simulate every other problem for which we can verify quickly that a solution is correct. In this sense, NP-complete problems are the hardest of the problems to which solutions can be verified quickly. If we could find solutions of some NP-complete problem quickly, we could quickly find the solutions of every other problem to which a given solution can be easily verified. The name "NP-complete" is short for "nondeterministic polynomial-time complete". In this name, "nondeterministic" refers to nondeterministic Turing machines, a way of mathematically formalizing the idea of a brute-force search algorithm. Polynomial time refers to an amount of time that is considered "quick" for a dete ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Naïve Algorithm
In mathematics and computer science, an algorithm () is a finite sequence of rigorous instructions, typically used to solve a class of specific problems or to perform a computation. Algorithms are used as specifications for performing calculations and data processing. More advanced algorithms can perform automated deductions (referred to as automated reasoning) and use mathematical and logical tests to divert the code execution through various routes (referred to as automated decision-making). Using human characteristics as descriptors of machines in metaphorical ways was already practiced by Alan Turing with terms such as "memory", "search" and "stimulus". In contrast, a heuristic is an approach to problem solving that may not be fully specified or may not guarantee correct or optimal results, especially in problem domains where there is no well-defined correct or optimal result. As an effective method, an algorithm can be expressed within a finite amount of space and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Big O Notation
Big ''O'' notation is a mathematical notation that describes the limiting behavior of a function when the argument tends towards a particular value or infinity. Big O is a member of a family of notations invented by Paul Bachmann, Edmund Landau, and others, collectively called Bachmann–Landau notation or asymptotic notation. The letter O was chosen by Bachmann to stand for '' Ordnung'', meaning the order of approximation. In computer science, big O notation is used to classify algorithms according to how their run time or space requirements grow as the input size grows. In analytic number theory, big O notation is often used to express a bound on the difference between an arithmetical function and a better understood approximation; a famous example of such a difference is the remainder term in the prime number theorem. Big O notation is also used in many other fields to provide similar estimates. Big O notation characterizes functions according to their growth rates: d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Substitution Matrix
In bioinformatics and evolutionary biology, a substitution matrix describes the frequency at which a character in a nucleotide sequence or a protein sequence changes to other character states over evolutionary time. The information is often in the form of log odds of finding two specific character states aligned and depends on the assumed number of evolutionary changes or sequence dissimilarity between compared sequences. It is an application of a stochastic matrix. Substitution matrices are usually seen in the context of amino acid or DNA sequence alignments, where they are used to calculate similarity scores between the aligned sequences. Background In the process of evolution, from one generation to the next the amino acid sequences of an organism's proteins are gradually altered through the action of DNA mutations. For example, the sequence ALEIRYLRD could mutate into the sequence ALEINYLRD in one step, and possibly AQEINYQRD over a longer period of evolutionar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gap Penalty
A Gap penalty is a method of scoring alignments of two or more sequences. When aligning sequences, introducing gaps in the sequences can allow an alignment algorithm to match more terms than a gap-less alignment can. However, minimizing gaps in an alignment is important to create a useful alignment. Too many gaps can cause an alignment to become meaningless. Gap penalties are used to adjust alignment scores based on the number and length of gaps. The five main types of gap penalties are constant, linear, affine, convex, and profile-based. Applications * Genetic sequence alignment - In bioinformatics, gaps are used to account for genetic mutations occurring from insertions or deletions in the sequence, sometimes referred to as ''indels''. Insertions or deletions can occur due to single mutations, unbalanced crossover in meiosis, slipped strand mispairing, and chromosomal translocation. The notion of a gap in an alignment is important in many biological applications, since the i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |