|
T4 RII System
The T4 ''r''II system is an experimental system developed in the 1950s by Seymour Benzer for studying the substructure of the gene. The experimental system is based on genetic crosses of different mutant strains of bacteriophage T4, a virus that infects the bacteria ''Escherichia coli''. Origin One type of mutation in the T4 bacteriophage identified by researchers in phage genetics by the 1950s was known as ''r'' (for ''rapid''), which caused the phage to destroy bacteria more quickly than normal. These could be spotted easily because they would produce larger plaques rather than the smaller plaques characteristic of the wild type virus. Through genetic mapping, the researchers had identified specific regions in the T4 chromosome, called the ''r''I, ''r''II, and ''r''III loci, associated with the ''r'' mutants. In 1952, while performing experiments with ''r''II mutants, Seymour Benzer found a strain that did not behave normally. By 1953, after the publication of Watson and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Experimental System
In scientific research, an experimental system is the physical, technical and procedural basis for an experiment or series of experiments. Historian of science Hans-Jörg Rheinberger defines an experimental system as: "A basic unit of experimental activity combining local, technical, instrumental, institutional, social, and epistemic aspects." Scientists (particularly laboratory biologists) and historians and philosophers of biology have pointed to the development and spread of successful experimental systems, such as those based on popular model organism or scientific apparatus, as key elements in the history of science, particularly since the early 20th century. The choice of an appropriate experimental system is often seen as critical for a scientist's long-term success, as experimental systems can be very productive for some kinds of questions and less productive for others, acquiring a sort of momentum that takes research in unpredicted directions. A successful experimental ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Max Delbrück
Max Ludwig Henning Delbrück (; September 4, 1906 – March 9, 1981) was a German–American biophysicist who participated in launching the molecular biology research program in the late 1930s. He stimulated physical science, physical scientists' interest into biology, especially as to basic research to physically explain genes, mysterious at the time. Formed in 1945 and led by Delbrück along with Salvador Luria and Alfred Hershey, the Phage Group made substantial headway unraveling important aspects of genetics. The three shared the 1969 Nobel Prize in Physiology or Medicine "for their discoveries concerning the replication mechanism and the genetic structure of viruses"."The Nobel Prize in Physiology or Medicine 1969" Nobel Media AB 2013, ''Nobelprize.org'', Web access November 6, 2013. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Recombination
Genetic recombination (also known as genetic reshuffling) is the exchange of genetic material between different organisms which leads to production of offspring with combinations of traits that differ from those found in either parent. In eukaryotes, genetic recombination during meiosis can lead to a novel set of genetic information that can be further passed on from parents to offspring. Most recombination occurs naturally and can be classified into two types: (1) ''interchromosomal'' recombination, occurring through independent assortment of alleles whose loci are on different but homologous chromosomes (random orientation of pairs of homologous chromosomes in meiosis I); & (2) ''intrachromosomal'' recombination, occurring through crossing over. During meiosis in eukaryotes, genetic recombination involves the pairing of homologous chromosomes. This may be followed by information transfer between the chromosomes. The information transfer may occur without physical exchange (a se ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Richard Feynman
Richard Phillips Feynman (; May 11, 1918 – February 15, 1988) was an American theoretical physicist, known for his work in the path integral formulation of quantum mechanics, the theory of quantum electrodynamics, the physics of the superfluidity of supercooled liquid helium, as well as his work in particle physics for which he proposed the parton model. For contributions to the development of quantum electrodynamics, Feynman received the Nobel Prize in Physics in 1965 jointly with Julian Schwinger and Shin'ichirō Tomonaga. Feynman developed a widely used pictorial representation scheme for the mathematical expressions describing the behavior of subatomic particles, which later became known as Feynman diagrams. During his lifetime, Feynman became one of the best-known scientists in the world. In a 1999 poll of 130 leading physicists worldwide by the British journal ''Physics World'', he was ranked the seventh-greatest physicist of all time. He assisted in the development o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
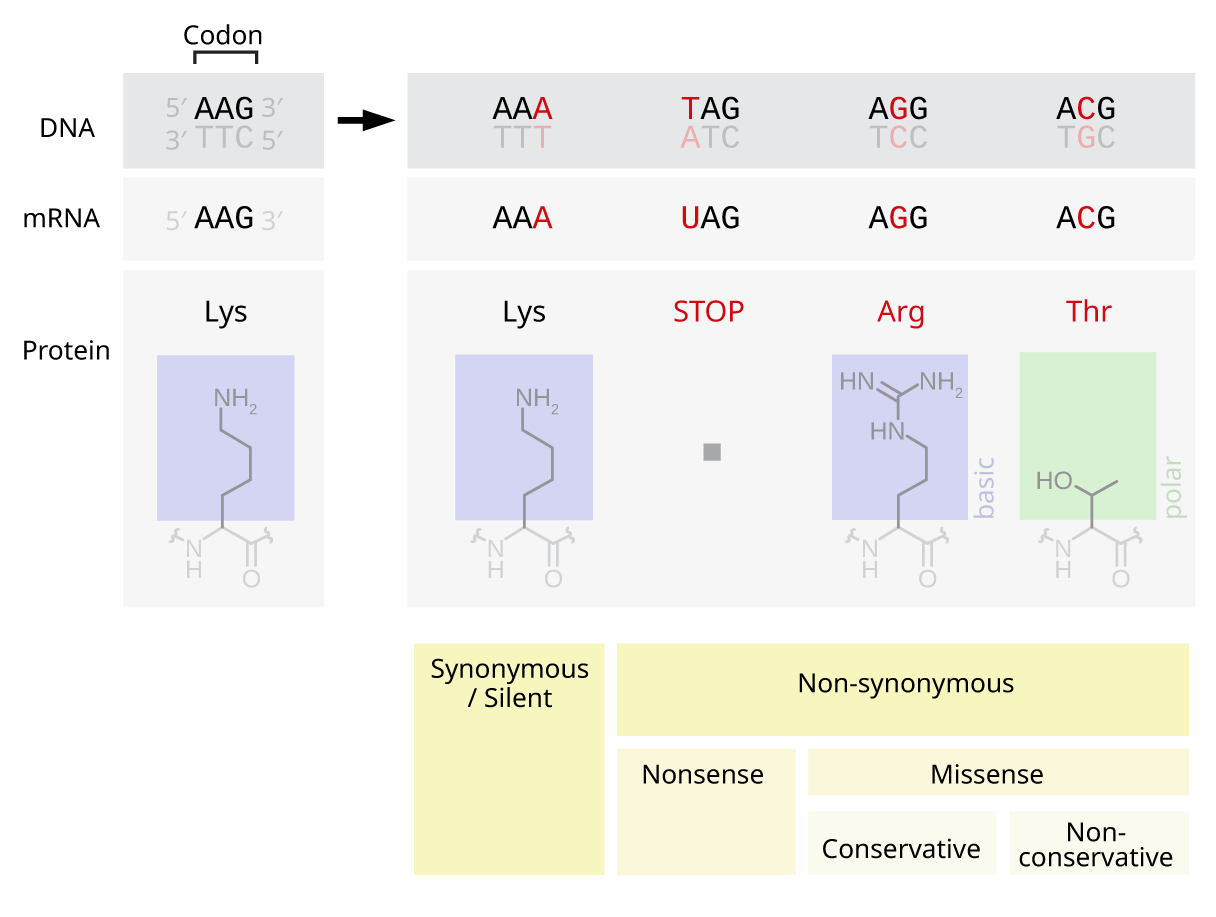
Genetic Code
The genetic code is the set of rules used by living cells to translate information encoded within genetic material ( DNA or RNA sequences of nucleotide triplets, or codons) into proteins. Translation is accomplished by the ribosome, which links proteinogenic amino acids in an order specified by messenger RNA (mRNA), using transfer RNA (tRNA) molecules to carry amino acids and to read the mRNA three nucleotides at a time. The genetic code is highly similar among all organisms and can be expressed in a simple table with 64 entries. The codons specify which amino acid will be added next during protein biosynthesis. With some exceptions, a three-nucleotide codon in a nucleic acid sequence specifies a single amino acid. The vast majority of genes are encoded with a single scheme (see the RNA codon table). That scheme is often referred to as the canonical or standard genetic code, or simply ''the'' genetic code, though variant codes (such as in mitochondria) exist. History Effor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nonsense Mutation
In genetics, a nonsense mutation is a point mutation in a sequence of DNA that results in a premature stop codon, or a ''nonsense codon'' in the transcribed mRNA, and in leading to a truncated, incomplete, and usually nonfunctional protein product. The functional effect of a nonsense mutation depends on the location of the stop codon within the coding DNA. For example, the effect of a nonsense mutation depends on the proximity of the nonsense mutation to the original stop codon, and the degree to which functional subdomains of the protein are affected. As nonsense mutations leads to premature termination of polypeptide chains; they are also called chain termination mutations. Missense mutations differ from nonsense mutations since they are point mutations that exhibit a single nucleotide change to cause substitution of a different amino acid. A nonsense mutation also differs from a nonstop mutation, which is a point mutation that removes a stop codon. About 10% of patients facin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Missense
In genetics, a missense mutation is a point mutation in which a single nucleotide change results in a codon that codes for a different amino acid. It is a type of nonsynonymous substitution. Substitution of protein from DNA mutations Missense mutation refers to a change in one amino acid in a protein, arising from a point mutation in a single nucleotide. Missense mutation is a type of nonsynonymous substitution in a DNA sequence. Two other types of nonsynonymous substitution are the nonsense mutations, in which a codon is changed to a premature stop codon that results in truncation of the resulting protein, and the nonstop mutations, in which a stop codon erasement results in a longer, nonfunctional protein. Missense mutations can render the resulting protein nonfunctional, and such mutations are responsible for human diseases such as Epidermolysis bullosa, sickle-cell disease, SOD1 mediated ALS, and a substantial number of cancers. In the most common variant of sickle-cell dis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Point Mutation
A point mutation is a genetic mutation where a single nucleotide base is changed, inserted or deleted from a DNA or RNA sequence of an organism's genome. Point mutations have a variety of effects on the downstream protein product—consequences that are moderately predictable based upon the specifics of the mutation. These consequences can range from no effect (e.g. synonymous mutations) to deleterious effects (e.g. frameshift mutations), with regard to protein production, composition, and function. Causes Point mutations usually take place during DNA replication. DNA replication occurs when one double-stranded DNA molecule creates two single strands of DNA, each of which is a template for the creation of the complementary strand. A single point mutation can change the whole DNA sequence. Changing one purine or pyrimidine may change the amino acid that the nucleotides code for. Point mutations may arise from spontaneous mutations that occur during DNA replication. The rate of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Deletion (genetics)
In genetics, a deletion (also called gene deletion, deficiency, or deletion mutation) (sign: Δ) is a mutation (a genetic aberration) in which a part of a chromosome or a sequence of DNA is left out during DNA replication. Any number of nucleotides can be deleted, from a single base to an entire piece of chromosome. Some chromosomes have fragile spots where breaks occur which result in the deletion of a part of chromosome. The breaks can be induced by heat, viruses, radiations, chemicals. When a chromosome breaks, a part of it is deleted or lost, the missing piece of chromosome is referred to as deletion or a deficiency. For synapsis to occur between a chromosome with a large intercalary deficiency and a normal complete homolog, the unpaired region of the normal homolog must loop out of the linear structure into a deletion or compensation loop. The smallest single base deletion mutations occur by a single base flipping in the template DNA, followed by template DNA strand sli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cistron
A cistron is an alternative term for "gene". The word cistron is used to emphasize that genes exhibit a specific behavior in a cis-trans test; distinct positions (or loci) within a genome are cistronic. History The words ''cistron'' and ''gene'' were coined before the advancing state of biology made it clear that the concepts they refer to are practically equivalent. The same historical naming practices are responsible for many of the synonyms in the life sciences. The term cistron was coined by Seymour Benzer in an article entitled ''The elementary units of heredity''. also reprinted in The cistron was defined by an operational test applicable to most organisms that is sometimes referred to as a cis-trans test, but more often as a complementation test. Definition For example, suppose a mutation at a chromosome position x is responsible for a change in recessive trait in a diploid organism (where chromosomes come in pairs). We say that the mutation is recessive because the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |