|
Schinkia
''Schinkia'' is a genus of Gram-positive rod-shaped bacteria in the family '' Bacillaceae'' from the order '' Bacillales.'' The type species of this genus is ''Schinkia azotoformans.'' Members of ''Schinkia'' are previously species belonging to '' Bacillus'', a genus that has been recognized as displaying extensive polyphyly and phylogenetic heterogeneity due to the vague criteria (such as the ability to form endospores in the presence of oxygen) previously used to assign species to this clade. Multiple studies using comparative phylogenetic analyses have been published in an attempt to clarify the evolutionary relationships between ''Bacillus'' species, resulting in the establishment of numerous novel genera such as '' Alkalihalobacillus'', ''Brevibacillus'', ''Solibacillus'', ''Alicyclobacillus'', ''Virgibacillus'', and Evansella. In addition, the genus ''Bacillus'' has been restricted to only include species closely related to '' Bacillus subtilis'' and '' Bacillus cereus.'' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacillus Azotoformans
''Bacillus azotoformans'' is a species of bacteria within the genus ''Bacillus ''Bacillus'' (Latin "stick") is a genus of Gram-positive, rod-shaped bacteria, a member of the phylum ''Bacillota'', with 266 named species. The term is also used to describe the shape (rod) of other so-shaped bacteria; and the plural ''Bacilli ...''. Novel nitrite reductases have been isolated from strains of this species. This species has been recently transferred into the genus '' Schinkia''. The correct nomenclature is ''Schinkia azotoformans.'' References External linksType strain of ''Bacillus azotoformans'' at Bac''Dive'' - the Bacterial Diversity Metadatabase azotoformans Bacteria described in 1983 {{bacilli-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacillaceae
The Bacillaceae are a family of gram-positive, heterotrophic, rod-shaped bacteria that may produce endospores. Motile members of this family are characterized by peritrichous flagella. Some Bacillaceae are aerobic, while others are facultative or strict anaerobes. Most are not pathogenic, but ''Bacillus'' species are known to cause disease in humans. Gram-variable cell wall Some Bacillaceae, such as the genera '' Filobacillus, Lentibacillus,'' and '' Halobacillus'', stain Gram-negative or Gram-variable, but are known to have a Gram-positive cell wall.Lim, J.M., Jeon, C.O., Song, S.M., and C.J. Kim. 2005''Pontibacillus chungwhensis gen. nov., sp. nov., a moderately halophilic Gram-positive bacterium from a solar saltern in Korea'' Int. J. Syst. Evol. Microbiol. 55:165-170. Nomenclature Taxa within this family are sometimes colloquially identified as bacilli. However, this term is ambiguous because it does not distinguish between class Bacilli, order Bacillales, family Bacilla ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria are vital in many stages of the nutrient cycle by recycling nutrients such as the fixation of nitrogen from the atmosphere. The nutrient cycle includes the decomposition of dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy. Bacteria also live in symbiotic and parasitic relationsh ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacillus Cereus
''Bacillus cereus'' is a Gram-positive rod-shaped bacterium commonly found in soil, food, and marine sponges. The specific name, ''cereus'', meaning "waxy" in Latin, refers to the appearance of colonies grown on blood agar. Some strains are harmful to humans and cause foodborne illness due to their spore-forming nature, while other strains can be beneficial as probiotics for animals, and even exhibit mutualism with certain plants. ''B. cereus'' bacteria may be anaerobes or facultative anaerobes, and like other members of the genus ''Bacillus'', can produce protective endospores. They have a wide range of virulence factors, including phospholipase C, cereulide, sphingomyelinase, metalloproteases, and cytotoxin K, many of which are regulated via quorum sensing. ''B. cereus'' strains exhibit flagellar motility. The ''Bacillus cereus'' group comprises seven closely related species: ''B. cereus'' ''sensu stricto'' (referred to herein as ''B. cereus''), '' B. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome Taxonomy Database
The Genome Taxonomy Database (GTDB) is an online database that maintains information on a proposed nomenclature of prokaryotes, following a phylogenomic approach based on a set of conserved single-copy proteins. In addition to breaking up paraphyletic groups, this method also reassigns taxonomic ranks algorithmically, creating new names in both cases. Information for archaea was added in 2020, along with a species classification based on average nucleotide identity. Each update incorporates new genomes as well as human adjustments to the taxonomy. An open-source tool called GTDB-Tk is available to classify draft genomes into the GTDB hierarchy. The GTDB system, via GTDB-Tk, has been used to catalogue not-yet-named bacteria in the human gut microbiome and other metagenomic sources. The GTDB is incorporated into the ''Bergey's Manual of Systematics of Archaea and Bacteria'' in 2019 as its phylogenomic resource. See also * PhyloCode * National Center for Biotechnology Informa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Monophyly
In cladistics for a group of organisms, monophyly is the condition of being a clade—that is, a group of taxa composed only of a common ancestor (or more precisely an ancestral population) and all of its lineal descendants. Monophyletic groups are typically characterised by shared derived characteristics ( synapomorphies), which distinguish organisms in the clade from other organisms. An equivalent term is holophyly. The word "mono-phyly" means "one-tribe" in Greek. Monophyly is contrasted with paraphyly and polyphyly as shown in the second diagram. A ''paraphyletic group'' consists of all of the descendants of a common ancestor minus one or more monophyletic groups. A '' polyphyletic group'' is characterized by convergent features or habits of scientific interest (for example, night-active primates, fruit trees, aquatic insects). The features by which a polyphyletic group is differentiated from others are not inherited from a common ancestor. These definitions have taken ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
UDP-N-acetylmuramoylalanyl-D-glutamate-2,6-diamino-pimelate Ligase
UDP-''N''-acetylmuramoyl-L-alanyl-D-glutamate—2,6-diaminopimelate ligase (, ''MurE synthetase'', ''UDP-N-acetylmuramoyl-L-alanyl-D-glutamate:meso-2,6-diamino-heptanedioate ligase (ADP-forming)'', ''UDP-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminopimelate synthetase'', ''UDP-N-acetylmuramoylalanyl-D-glutamate—2,6-diaminopimelate ligase'') is an enzyme with systematic name ''UDP-N-acetylmuramoyl-L-alanyl-D-glutamate:meso-2,6-diaminoheptanedioate gamma-ligase (ADP-forming)''. This enzyme catalyses the following chemical reaction : ATP + UDP-N-acetylmuramoyl-L-alanyl-D-glutamate + meso-2,6-diaminoheptanedioate \rightleftharpoons ADP + phosphate + UDP-N-acetylmuramoyl-L-alanyl-D-gamma-glutamyl-meso-2,6-diaminoheptanedioate This enzyme takes part in synthesis of a cell-wall peptide. References External links * {{Portal bar, Biology, border=no EC 6.3.2 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Acetolactate Synthase
The acetolactate synthase (ALS) enzyme (also known as acetohydroxy acid or acetohydroxyacid synthase, abbr. AHAS) is a protein found in plants and micro-organisms. ALS catalyzes the first step in the synthesis of the branched-chain amino acids (valine, leucine, and isoleucine). A human protein of yet unknown function, sharing some sequence similarity with bacterial ALS, is encoded by the ILVBL (ilvB-like) gene. Structure Gene Human ILVBL gene has 17 exons resides on chromosome 19 at q13.1. Protein The catalytic peptide of ALS in '' Arabidopsis thaliana'' (mouse-eared cress) is a chloroplastic protein consisting of 670 residues, the last 615 of which form the active form. Three main domains are found, with two thiamine pyrophosphate sandwiching a DHS-like NAD/FAD-binding domain. In SCOP assignment, these subunits are named d1yhya1, d1yhya2, and d1yhya3 from the N-terminal to the C-termianl. The structure of acetolactate synthase that was used for the picture on this page ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydroxymethylbilane Synthase
Porphobilinogen deaminase (hydroxymethylbilane synthase, or uroporphyrinogen I synthase) is an enzyme () that in humans is encoded by the HMBS gene. Porphobilinogen deaminase is involved in the third step of the heme biosynthetic pathway. It catalyzes the head to tail condensation of four porphobilinogen molecules into the linear hydroxymethylbilane while releasing four ammonia molecules: :4 porphobilinogen + H2O \rightleftharpoons hydroxymethylbilane + 4 NH3 Structure and function Functionally, porphobilinogen deaminase catalyzes the loss of ammonia from the porphobilinogen monomer (deamination) and its subsequent polymerization to a linear tetrapyrrole, which is released as hydroxymethylbilane: The structure of 40-42 kDa porphobilinogen deaminase, which is highly conserved amongst organisms, consists of three domains. Domains 1 and 2 are structurally very similar: each consisting of five beta-sheets and three alpha helices in humans. Domain 3 is positioned between the other ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Penicillin Binding Protein 2A
''mecA'' is a gene found in bacterial cells which allows them to be resistant to antibiotics such as methicillin, penicillin and other penicillin-like antibiotics. The bacteria strain most commonly known to carry ''mecA'' is methicillin-resistant ''Staphylococcus aureus'' (MRSA). In '' Staphylococcus'' species, ''mecA'' is spread through the staphylococcal chromosome cassette SCC''mec'' genetic element. Resistant strains cause many hospital-acquired infections. ''mecA'' encodes the protein PBP2A (penicillin-binding protein 2A), a transpeptidase that helps form the bacterial cell wall. PBP2A has a lower affinity for beta-lactam antibiotics such as methicillin and penicillin than DD-transpeptidase does, so it does not bind to the ringlike structure of penicillin-like antibiotics. This enables transpeptidase activity in the presence of beta-lactams, preventing them from inhibiting cell wall synthesis. The bacteria can then replicate as normal. History Methicillin resistance f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
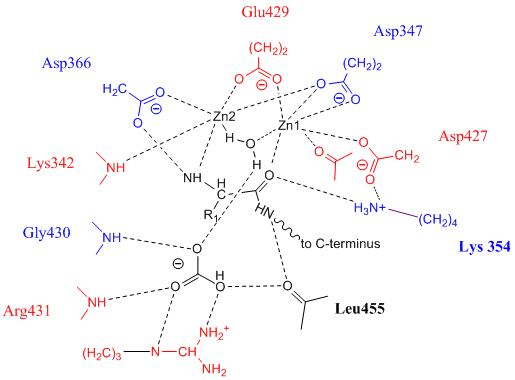
Leucyl Aminopeptidase
Leucyl aminopeptidases (, ''leucine aminopeptidase'', ''LAPs'', ''leucyl peptidase'', ''peptidase S'', ''cytosol aminopeptidase'', ''cathepsin III'', ''L-leucine aminopeptidase'', ''leucinaminopeptidase'', ''leucinamide aminopeptidase'', ''FTBL proteins'', ''proteinates FTBL'', ''aminopeptidase II'', ''aminopeptidase III'', ''aminopeptidase I'') are enzymes that preferentially catalyze the hydrolysis of leucine residues at the N-terminus of peptides and proteins. Other N-terminal residues can also be cleaved, however. LAPs have been found across superkingdoms. Identified LAPs include human LAP, bovine lens LAP, porcine LAP, ''Escherichia coli'' (''E. coli'') LAP (also known as PepA or XerB), and the solanaceous-specific acidic LAP (LAP-A) in tomato (''Solanum lycopersicum''). Enzyme description, structure, and active site The active sites in PepA and in bovine lens LAP have been found to be similar. Shown in the picture below is the proposed model for the active site of LAP-A in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |