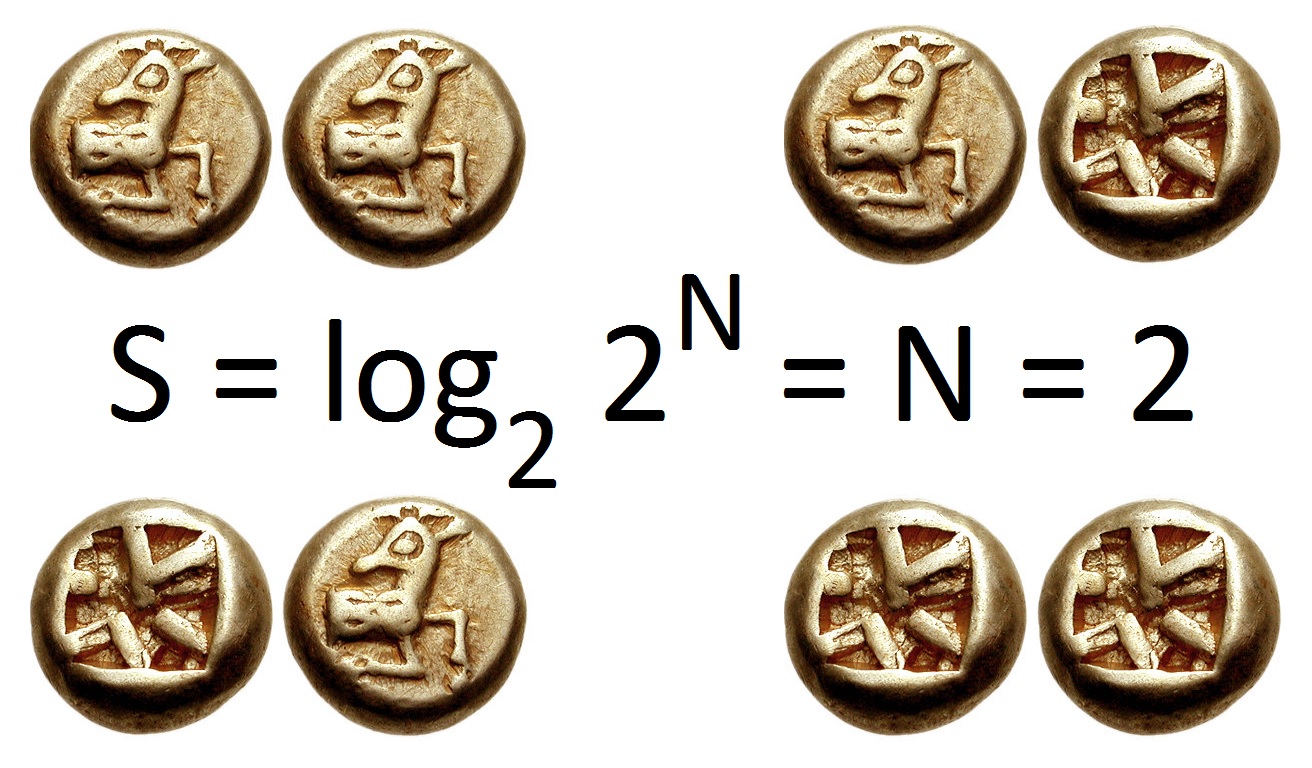|
NeuroKit
NeuroKit (''"nk"'') is an open source toolbox for physiological signal processing. The most recent version, ''NeuroKit2'', is written in Python and is available from the PyPI package repository. As of June 2022, the software was used in 94 scientific publications. NeuroKit2 is presented as one of the most popular and contributor-friendly open-source software for neurophysiology based on the number of downloads, the number of contributors, and other GitHub metrics. History The first version of ''NeuroKit'' was created as a PhD side-project of Dominique Makowski in 2017. It was officially deprecated in 2020 and has been replaced by the current version, ''NeuroKit2''. A few major updates have been released since: * February 08, 2021: The 0.1.0 release coincides with the first publication of the software. * May 18, 2022: The 0.2.0 release coincides with an overhaul of the documentation. Features NeuroKit2 includes tools to work with cardiac activity from electrocardiography (ECG ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fractal Physiology
Fractal physiology refers to the study of physiological systems using complexity science methods, such as chaos measure, entropy, and fractal dimensions. The underlying assumption is that biological systems are complex and exhibit non-linear patterns of activity, and that characterizing that complexity (using dedicated mathematical approaches) is useful to understand, and make inferences and predictions about the system. Main Findings Neurophysiology Quantifications of the complexity of brain activity is used in the context of neuropsychiatric diseases and mental states characterization, such as schizophrenia, affective disorders, or neurodegenerative disorders. Particularly, diminished EEG complexity is typically associated with increased symptomatology. Cardiovascular systems The complexity of Heart Rate Variability is a useful predictor of cardiovascular health. Software In Python, NeuroKit provides a comprehensive set of functions for complexity analysis of physiological ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Neuroimaging Software
Neuroimaging software is used to study the structure and function of the brain. To see an NIH Blueprint for Neuroscience Research funded clearinghouse of many of these software applications, as well as hardware, etc. go to the NITRC web site. * 3D Slicer Extensible, free open source multi-purpose software for visualization and analysis. * Amira 3D visualization and analysis software * Analysis of Functional NeuroImages (AFNI) * Analyze developed by the Biomedical Imaging Resource (BIR) at Mayo Clinic. * Brain Image Analysis Package * CamBA * Caret Van Essen Lab, Washington University in St. Louis * CONN (functional connectivity toolbox) Diffusion Imaging in Python (DIPY)DL+DiReCT* EEGLAB * FMRIB Software Library (FSL) * FreeSurfer * Imarisbr>Imaris for Neuroscientists* ISAS (Ictal-Interictal SPECT Analysis by SPM) * LONI Pipeline, Laboratory of Neuro Imaging, USC * Mango * NITRC The Neuroimaging Informatics Tools and Resources Clearinghouse. An NIH funded database o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MNE-Python
MNE-Python (''"MNE"'') is an open source toolbox for EEG and MEG signal processing. It is written in Python and is available from the PyPI package repository. See also * Neurophysiological Biomarker Toolbox ''(MatLab)'' * EEGLAB ''(MatLab)'' * NeuroKit NeuroKit (''"nk"'') is an open source toolbox for physiological signal processing. The most recent version, ''NeuroKit2'', is written in Python and is available from the PyPI package repository. As of June 2022, the software was used in 94 scient ... ''(Python)'' References {{science-software-stub Python (programming language) scientific libraries Neuroimaging software ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EEGLAB
EEGLAB is a MATLAB toolbox distributed under the free BSD license for processing data from electroencephalography (EEG), magnetoencephalography (MEG), and other electrophysiological signals. Along with all the basic processing tools, EEGLAB implements independent component analysis (ICA), time/frequency analysis, artifact rejection, and several modes of data visualization. EEGLAB allows users to import their electrophysiological data in about 20 binary file formats, preprocess the data, visualize activity in single trials, and perform ICA. Artifactual ICA components may be subtracted from the data. Alternatively, ICA components representing brain activity may be further processed and analyzed. EEGLAB also allows users to group data from several subjects, and to cluster their independent components. History In 1997, a set of data processing functions was first released on the Internet by Scott Makeig in the Computational Neurobiology Laboratory directed by Terry Sejnowski at the Salk ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Neurophysiological Biomarker Toolbox
The Neurophysiological Biomarker Toolbox (NBT) is an open source MATLAB toolbox for the computation and integration of neurophysiological biomarkers (e.g., biomarkers based on EEG or MEG recordings). The NBT toolbox has so far been used in seven peer-reviewed research articles, and has a broad user base of more than 1000 users. The NBT toolbox provides unique features for analysis of resting-state EEG or MEG recordings. NBT offers a pipeline from data storage to statistics including artifact rejection, signal visualization, biomarker computation, statistical testing, and biomarker databasing. NBT allows for easy implementation of new biomarkers, and incorporates an online wiki (the NBTwiki) that aims at facilitating collaboration among NBT users including extensive help and tutorials. The standardised way of data storage and analysis that NBT proposes allow different research projects to merge, compare, or share their data and biomarker algorithms. Features Neuronal oscilla ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Respiratory Variability
The respiratory system (also respiratory apparatus, ventilatory system) is a biological system consisting of specific organs and structures used for gas exchange in animals and plants. The anatomy and physiology that make this happen varies greatly, depending on the size of the organism, the environment in which it lives and its evolutionary history. In land animals the respiratory surface is internalized as linings of the lungs. Gas exchange in the lungs occurs in millions of small air sacs; in mammals and reptiles these are called alveoli, and in birds they are known as atria. These microscopic air sacs have a very rich blood supply, thus bringing the air into close contact with the blood. These air sacs communicate with the external environment via a system of airways, or hollow tubes, of which the largest is the trachea, which branches in the middle of the chest into the two main bronchi. These enter the lungs where they branch into progressively narrower secondary and terti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Python (programming Language) Scientific Libraries
Python may refer to: Snakes * Pythonidae, a family of nonvenomous snakes found in Africa, Asia, and Australia ** ''Python'' (genus), a genus of Pythonidae found in Africa and Asia * Python (mythology), a mythical serpent Computing * Python (programming language), a widely used programming language * Python, a native code compiler for CMU Common Lisp * Python, the internal project name for the PERQ 3 computer workstation People * Python of Aenus (4th-century BCE), student of Plato * Python (painter), (ca. 360–320 BCE) vase painter in Poseidonia * Python of Byzantium, orator, diplomat of Philip II of Macedon * Python of Catana, poet who accompanied Alexander the Great * Python Anghelo (1954–2014) Romanian graphic artist Roller coasters * Python (Efteling), a roller coaster in the Netherlands * Python (Busch Gardens Tampa Bay), a defunct roller coaster * Python (Coney Island, Cincinnati, Ohio), a steel roller coaster Vehicles * Python (automobile maker), an Australian car co ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fractal Dimensions
In mathematics, more specifically in fractal geometry, a fractal dimension is a ratio providing a statistical index of complexity comparing how detail in a pattern (strictly speaking, a fractal pattern) changes with the Scaling (geometry), scale at which it is measured. It has also been characterized as a measure of the Space-filling curve, space-filling capacity of a pattern that tells how a fractal scales differently from the space it is embedded in; a fractal dimension does not have to be an integer. The essential idea of "fractured" Hausdorff dimension, dimensions has a long history in mathematics, but the term itself was brought to the fore by Benoit Mandelbrot based on How Long Is the Coast of Britain? Statistical Self-Similarity and Fractional Dimension, his 1967 paper on self-similarity in which he discussed ''fractional dimensions''. In that paper, Mandelbrot cited previous work by Lewis Fry Richardson describing the counter-intuitive notion that a coastline's measured le ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Entropy (information Theory)
In information theory, the entropy of a random variable is the average level of "information", "surprise", or "uncertainty" inherent to the variable's possible outcomes. Given a discrete random variable X, which takes values in the alphabet \mathcal and is distributed according to p: \mathcal\to , 1/math>: \Eta(X) := -\sum_ p(x) \log p(x) = \mathbb \log p(X), where \Sigma denotes the sum over the variable's possible values. The choice of base for \log, the logarithm, varies for different applications. Base 2 gives the unit of bits (or " shannons"), while base ''e'' gives "natural units" nat, and base 10 gives units of "dits", "bans", or " hartleys". An equivalent definition of entropy is the expected value of the self-information of a variable. The concept of information entropy was introduced by Claude Shannon in his 1948 paper "A Mathematical Theory of Communication",PDF archived froherePDF archived frohere and is also referred to as Shannon entropy. Shannon's theory defi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Complex System
A complex system is a system composed of many components which may interact with each other. Examples of complex systems are Earth's global climate, organisms, the human brain, infrastructure such as power grid, transportation or communication systems, complex software and electronic systems, social and economic organizations (like cities), an ecosystem, a living cell, and ultimately the entire universe. Complex systems are systems whose behavior is intrinsically difficult to model due to the dependencies, competitions, relationships, or other types of interactions between their parts or between a given system and its environment. Systems that are "complex" have distinct properties that arise from these relationships, such as nonlinearity, emergence, spontaneous order, adaptation, and feedback loops, among others. Because such systems appear in a wide variety of fields, the commonalities among them have become the topic of their independent area of research. In many cases, i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EEG Analysis
EEG analysis is exploiting mathematical signal analysis methods and computer technology to extract information from electroencephalography (EEG) signals. The targets of EEG analysis are to help researchers gain a better understanding of the brain; assist physicians in diagnosis and treatment choices; and to boost brain-computer interface (BCI) technology. There are many ways to roughly categorize EEG analysis methods. If a mathematical model is exploited to fit the sampled EEG signals, the method can be categorized as parametric, otherwise, it is a non-parametric method. Traditionally, most EEG analysis methods fall into four categories: time domain, frequency domain, time-frequency domain, and nonlinear methods. There are also later methods including deep neural networks (DNNs). Methods Frequency domain methods Frequency domain analysis, also known as spectral analysis, is the most conventional yet one of the most powerful and standard methods for EEG analysis. It gives ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EEG Microstates
EEG microstates are transient, patterned, quasi-stable states or patterns of an electroencephalography, electroencephalogram. These tend to last anywhere from milliseconds to seconds and are hypothesized to be the most basic instantiations of human neurology, neurological tasks, and are thus nicknamed "the atoms of thought". Microstate estimation and analysis was originally done using Alpha wave, alpha band activity, though broader bandwidth EEG bands are now typically used. The quasi-stability of microstates means that the "global [EEG] topography is fixed, but strength might vary and polarity invert." History The concept of temporal microstates of brain electrical activity during no-task resting and task execution (event-related microstates) was developed by Dietrich Lehmann and his collaborators (The KEY Institute for Brain-Mind Research, University of Zurich, Switzerland) between 1971 and 1987,( see ) Drs. Thomas Koenig (University Hospital of Psychiatry, Switzerland) and Die ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |