|
Neurophysiological Biomarker Toolbox
The Neurophysiological Biomarker Toolbox (NBT) is an open source MATLAB toolbox for the computation and integration of neurophysiological biomarkers (e.g., biomarkers based on EEG or MEG recordings). The NBT toolbox has so far been used in seven peer-reviewed research articles, and has a broad user base of more than 1000 users. The NBT toolbox provides unique features for analysis of resting-state EEG or MEG recordings. NBT offers a pipeline from data storage to statistics including artifact rejection, signal visualization, biomarker computation, statistical testing, and biomarker databasing. NBT allows for easy implementation of new biomarkers, and incorporates an online wiki (the NBTwiki) that aims at facilitating collaboration among NBT users including extensive help and tutorials. The standardised way of data storage and analysis that NBT proposes allow different research projects to merge, compare, or share their data and biomarker algorithms. Features Neuronal oscil ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Matlab
MATLAB (an abbreviation of "MATrix LABoratory") is a proprietary multi-paradigm programming language and numeric computing environment developed by MathWorks. MATLAB allows matrix manipulations, plotting of functions and data, implementation of algorithms, creation of user interfaces, and interfacing with programs written in other languages. Although MATLAB is intended primarily for numeric computing, an optional toolbox uses the MuPAD symbolic engine allowing access to symbolic computing abilities. An additional package, Simulink, adds graphical multi-domain simulation and model-based design for dynamic and embedded systems. , MATLAB has more than four million users worldwide. They come from various backgrounds of engineering, science, and economics. , more than 5000 global colleges and universities use MATLAB to support instruction and research. History Origins MATLAB was invented by mathematician and computer programmer Cleve Moler. The idea for MATLAB was base ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Biomarkers
In biomedical contexts, a biomarker, or biological marker, is a measurable indicator of some biological state or condition. Biomarkers are often measured and evaluated using blood, urine, or soft tissues to examine normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention. as cited in Biomarkers are used in many scientific fields. Medicine Biomarkers used in the medical field, are a part of a relatively new clinical toolset categorized by their clinical applications. The four main classes are molecular, physiologic, histologic and radiographic biomarkers. All four types of biomarkers have a clinical role in narrowing or guiding treatment decisions and follow a sub-categorization of being either predictive, prognostic, or diagnostic. Predictive Predictive molecular, cellular, or imaging biomarkers that pass validation can serve as a method of predicting clinical outcomes. Predictive biomarkers are used to help optimize ide ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
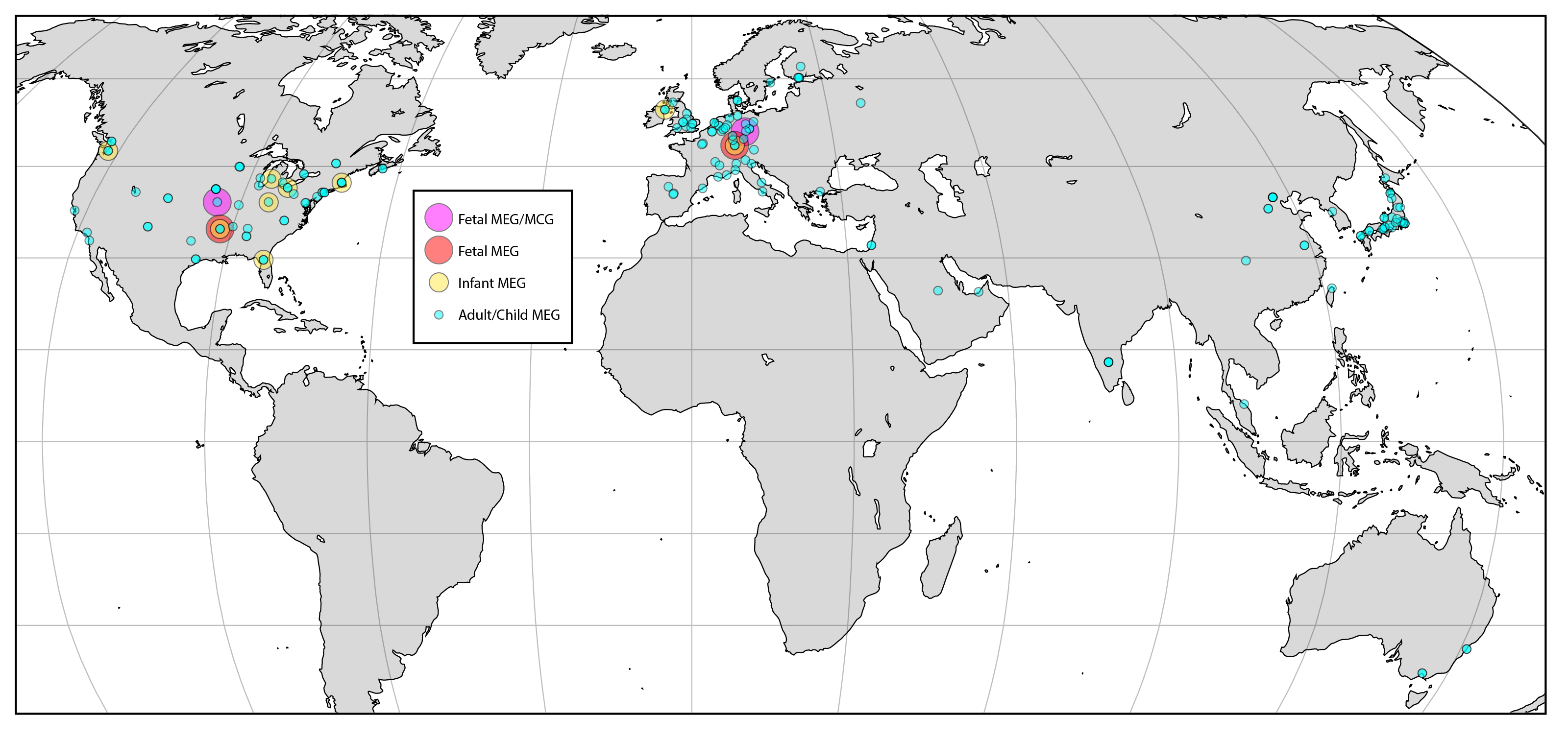
Magnetoencephalography
Magnetoencephalography (MEG) is a functional neuroimaging technique for mapping brain activity by recording magnetic fields produced by electric current, electrical currents occurring naturally in the human brain, brain, using very sensitive magnetometers. Arrays of SQUIDs (superconducting quantum interference devices) are currently the most common magnetometer, while the SERF (spin exchange relaxation-free) magnetometer is being investigated for future machines. Applications of MEG include basic research into perceptual and cognitive brain processes, localizing regions affected by pathology before surgical removal, determining the function of various parts of the brain, and neurofeedback. This can be applied in a clinical setting to find locations of abnormalities as well as in an experimental setting to simply measure brain activity. History MEG signals were first measured by University of Illinois physicist David Cohen (physicist), David Cohen in 1968, before the availabili ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Detrended Fluctuation Analysis
In stochastic processes, chaos theory and time series analysis, detrended fluctuation analysis (DFA) is a method for determining the statistical self-affinity of a signal. It is useful for analysing time series that appear to be long-memory processes (diverging correlation time, e.g. power-law decaying autocorrelation function) or 1/f noise. The obtained exponent is similar to the Hurst exponent, except that DFA may also be applied to signals whose underlying statistics (such as mean and variance) or dynamics are non-stationary (changing with time). It is related to measures based upon spectral techniques such as autocorrelation and Fourier transform. Peng et al. introduced DFA in 1994 in a paper that has been cited over 3,000 times as of 2022 and represents an extension of the (ordinary) fluctuation analysis (FA), which is affected by non-stationarities. Systematic studies of the advantages and limitations of the DFA method were performed by PCh Ivanov et al. in a serie ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EEGLAB
EEGLAB is a MATLAB toolbox distributed under the free BSD license for processing data from electroencephalography (EEG), magnetoencephalography (MEG), and other electrophysiological signals. Along with all the basic processing tools, EEGLAB implements independent component analysis (ICA), time/frequency analysis, artifact rejection, and several modes of data visualization. EEGLAB allows users to import their electrophysiological data in about 20 binary file formats, preprocess the data, visualize activity in single trials, and perform ICA. Artifactual ICA components may be subtracted from the data. Alternatively, ICA components representing brain activity may be further processed and analyzed. EEGLAB also allows users to group data from several subjects, and to cluster their independent components. History In 1997, a set of data processing functions was first released on the Internet by Scott Makeig in the Computational Neurobiology Laboratory directed by Terry Sejnowski at the Sal ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MNE-Python
MNE-Python (''"MNE"'') is an open source toolbox for EEG and MEG signal processing. It is written in Python and is available from the PyPI package repository. See also * Neurophysiological Biomarker Toolbox ''(MatLab)'' * EEGLAB ''(MatLab)'' * NeuroKit NeuroKit (''"nk"'') is an open source toolbox for physiology, physiological signal processing. The most recent version, ''NeuroKit2'', is written in Python_(programming_language), Python and is available from the Python Package Index, PyPI package ... ''(Python)'' References {{science-software-stub Python (programming language) scientific libraries Neuroimaging software ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NeuroKit
NeuroKit (''"nk"'') is an open source toolbox for physiology, physiological signal processing. The most recent version, ''NeuroKit2'', is written in Python_(programming_language), Python and is available from the Python Package Index, PyPI package repository. As of June 2022, the software was used in 94 scientific publications. NeuroKit2 is presented as one of the most popular and contributor-friendly open-source software for neurophysiology based on the number of downloads, the number of contributors, and other GitHub metrics. History The first version of ''NeuroKit'' was created as a PhD side-project of Dominique Makowski in 2017. It was officially deprecated in 2020 and has been replaced by the current version, ''NeuroKit2''. A few major updates have been released since: * February 08, 2021: The 0.1.0 release coincides with the first publication of the software. * May 18, 2022: The 0.2.0 release coincides with an overhaul of the documentation. ''NeuroKit'' has received the 20 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Free Statistical Software
Free statistical software is a practical alternative to commercial packages. Many of the free to use programs aim to be similar in function to commercial packages, in that they are general statistical packages that perform a variety of statistical analyses. Many other free to use programs were designed specifically for particular functions, like factor analysis, power analysis in sample size calculations, classification and regression trees, or analysis of missing data. Many of the free to use packages are fairly easy to learn, using menu systems. Many others are command-driven. Still others are meta-packages or statistical computing environments, which allow the user to code completely new statistical procedures. These packages come from a variety of sources, including governments, universities, and private individuals. This article is primarily a review of the general statistical packages. Brief history of free statistical software SAS (software) was among the first commerci ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Free Data Analysis Software
Free may refer to: Concept * Freedom, the ability to act or change without constraint or restriction * Emancipate, attaining civil and political rights or equality * Free (gratis), Free (''gratis''), free of charge * Gratis versus libre, the difference between the two common meanings of the adjective "free". Computing * Free (programming), a function that releases dynamically allocated memory for reuse * Free software, software usable and distributable with few restrictions and no payment *, an emoji in the Enclosed Alphanumeric Supplement block. Mathematics * Free object ** Free abelian group ** Free algebra ** Free group ** Free module ** Free semigroup * Free variable People * Free (surname) * Free (rapper) (born 1968), or Free Marie, American rapper and media personality * Free, a pseudonym for the activist and writer Abbie Hoffman * Free (active 2003–), American musician in the band FreeSol Arts and media Film and television * Free (film), ''Free'' (film), a 200 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Electroencephalography
Electroencephalography (EEG) is a method to record an electrogram of the spontaneous electrical activity of the brain. The biosignal, bio signals detected by EEG have been shown to represent the postsynaptic potentials of pyramidal neurons in the neocortex and allocortex. It is typically non-invasive, with the EEG electrodes placed along the scalp (commonly called "scalp EEG") using the 10–20 system (EEG), International 10–20 system, or variations of it. Electrocorticography, involving surgical placement of electrodes, is sometimes called Electrocorticography, "intracranial EEG". Clinical interpretation of EEG recordings is most often performed by visual inspection of the tracing or quantitative EEG, quantitative EEG analysis. Voltage fluctuations measured by the EEG bioamplifier, bio amplifier and electrodes allow the evaluation of normal Brain activity and meditation, brain activity. As the electrical activity monitored by EEG originates in neurons in the underlying Huma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |