|
EEGLAB
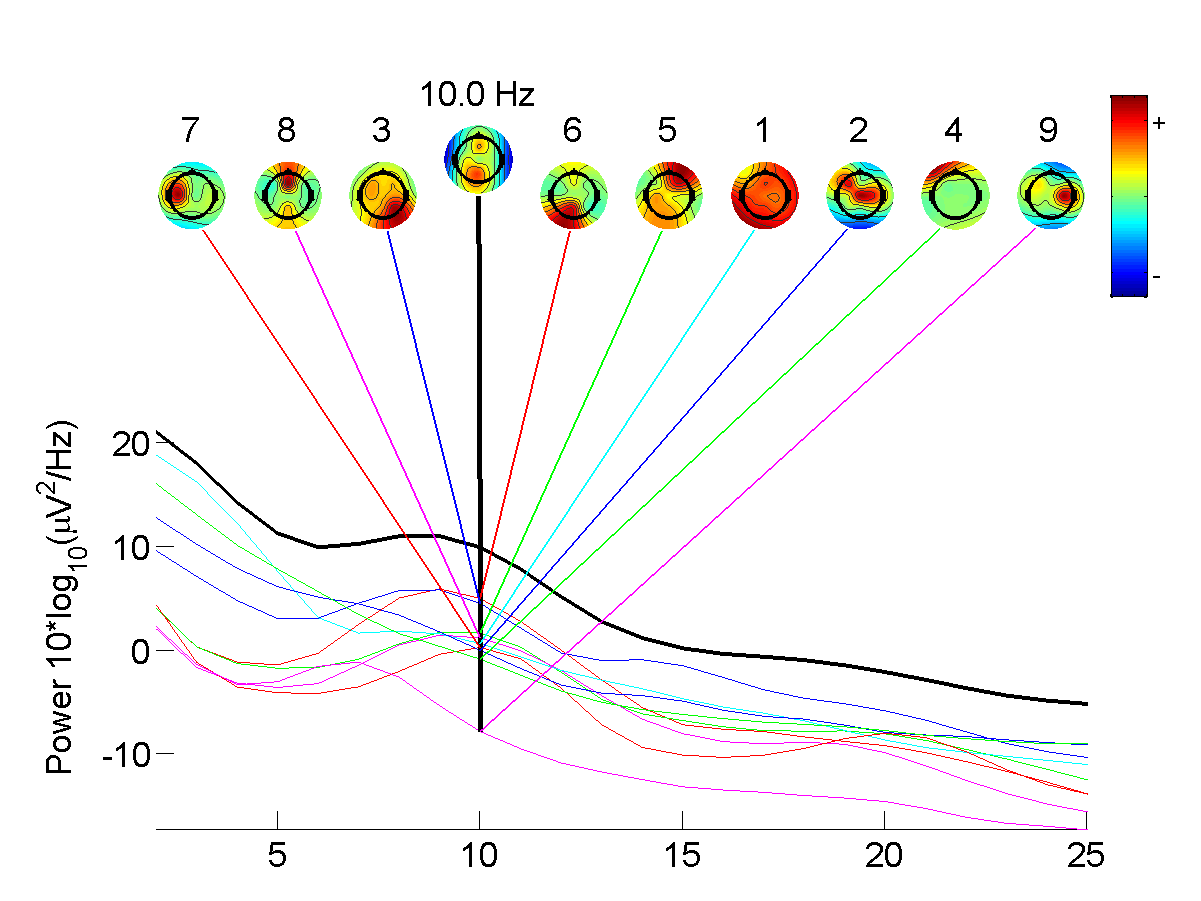
EEGLAB is a MATLAB toolbox distributed under the free BSD license for processing data from electroencephalography (EEG), magnetoencephalography (MEG), and other electrophysiological signals. Along with all the basic processing tools, EEGLAB implements independent component analysis (ICA), time/frequency analysis, artifact rejection, and several modes of data visualization. EEGLAB allows users to import their electrophysiological data in about 20 binary file formats, preprocess the data, visualize activity in single trials, and perform ICA. Artifactual ICA components may be subtracted from the data. Alternatively, ICA components representing brain activity may be further processed and analyzed. EEGLAB also allows users to group data from several subjects, and to cluster their independent components. History In 1997, a set of data processing functions was first released on the Internet by Scott Makeig in the Computational Neurobiology Laboratory directed by Terry Sejnowski at the Salk ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Independent Component Analysis
In signal processing, independent component analysis (ICA) is a computational method for separating a multivariate signal into additive subcomponents. This is done by assuming that at most one subcomponent is Gaussian and that the subcomponents are statistically independent from each other. ICA is a special case of blind source separation. A common example application is the "cocktail party problem" of listening in on one person's speech in a noisy room. Introduction Independent component analysis attempts to decompose a multivariate signal into independent non-Gaussian signals. As an example, sound is usually a signal that is composed of the numerical addition, at each time t, of signals from several sources. The question then is whether it is possible to separate these contributing sources from the observed total signal. When the statistical independence assumption is correct, blind ICA separation of a mixed signal gives very good results. It is also used for signals that are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Electroencephalography
Electroencephalography (EEG) is a method to record an electrogram of the spontaneous electrical activity of the brain. The biosignals detected by EEG have been shown to represent the postsynaptic potentials of pyramidal neurons in the neocortex and allocortex. It is typically non-invasive, with the EEG electrodes placed along the scalp (commonly called "scalp EEG") using the International 10-20 system, or variations of it. Electrocorticography, involving surgical placement of electrodes, is sometimes called " intracranial EEG". Clinical interpretation of EEG recordings is most often performed by visual inspection of the tracing or quantitative EEG analysis. Voltage fluctuations measured by the EEG bioamplifier and electrodes allow the evaluation of normal brain activity. As the electrical activity monitored by EEG originates in neurons in the underlying brain tissue, the recordings made by the electrodes on the surface of the scalp vary in accordance with their orientation and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Neurophysiological Biomarker Toolbox
The Neurophysiological Biomarker Toolbox (NBT) is an open source MATLAB toolbox for the computation and integration of neurophysiological biomarkers (e.g., biomarkers based on EEG or MEG recordings). The NBT toolbox has so far been used in seven peer-reviewed research articles, and has a broad user base of more than 1000 users. The NBT toolbox provides unique features for analysis of resting-state EEG or MEG recordings. NBT offers a pipeline from data storage to statistics including artifact rejection, signal visualization, biomarker computation, statistical testing, and biomarker databasing. NBT allows for easy implementation of new biomarkers, and incorporates an online wiki (the NBTwiki) that aims at facilitating collaboration among NBT users including extensive help and tutorials. The standardised way of data storage and analysis that NBT proposes allow different research projects to merge, compare, or share their data and biomarker algorithms. Features Neuronal oscilla ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Terry Sejnowski
Terrence Joseph Sejnowski (born 13 August 1947) is the Francis Crick Professor at the Salk Institute for Biological Studies where he directs the Computational Neurobiology Laboratory and is the director of the Crick-Jacobs center for theoretical and computational biology. He has performed pioneering research in neural networks and computational neuroscience. Sejnowski is also Professor of Biological Sciences and adjunct professor in the departments of neurosciences, psychology, cognitive science, computer science and engineering at the University of California, San Diego, where he is co-director of the Institute for Neural Computation. With Barbara Oakley, he co-created and taught ''Learning How To Learn: Powerful mental tools to help you master tough subjects'', the world's most popular online course, available on Coursera. Education and early life Born in Cleveland in 1947, Sejnowski received his B.S. in physics in 1968 from the Case Western Reserve University, M.A. in phy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Arnaud Delorme
Arnaud Delorme is a university professor at Paul Sabatier University in Toulouse, an adjunct faculty member at the Swartz Center for Computational Neuroscience at University of California, San Diego, a consulting research scientist at the Institute of Noetic Sciences. At UCSD, Dr. Delorme contributed to development of the widely used Matlab toolbox for electroencephalography (EEG) analysis, EEGLAB. He has been acknowledged for his contribution to the field of EEG research by being awarded a Bettencourt-Schueller young investigator award in 2002 and one of three of the ANT EEG Company 10-year Anniversary Young Researcher Awards in 2006. His research has focused on pure neuroscience Neuroscience is the science, scientific study of the nervous system (the brain, spinal cord, and peripheral nervous system), its functions and disorders. It is a Multidisciplinary approach, multidisciplinary science that combines physiology, an ... methods, as well as on the neuroscience of mind wand ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MNE-Python
MNE-Python (''"MNE"'') is an open source toolbox for EEG and MEG signal processing. It is written in Python and is available from the PyPI package repository. See also * Neurophysiological Biomarker Toolbox ''(MatLab)'' * EEGLAB ''(MatLab)'' * NeuroKit NeuroKit (''"nk"'') is an open source toolbox for physiological signal processing. The most recent version, ''NeuroKit2'', is written in Python and is available from the PyPI package repository. As of June 2022, the software was used in 94 scient ... ''(Python)'' References {{science-software-stub Python (programming language) scientific libraries Neuroimaging software ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
NeuroKit
NeuroKit (''"nk"'') is an open source toolbox for physiological signal processing. The most recent version, ''NeuroKit2'', is written in Python and is available from the PyPI package repository. As of June 2022, the software was used in 94 scientific publications. NeuroKit2 is presented as one of the most popular and contributor-friendly open-source software for neurophysiology based on the number of downloads, the number of contributors, and other GitHub metrics. History The first version of ''NeuroKit'' was created as a PhD side-project of Dominique Makowski in 2017. It was officially deprecated in 2020 and has been replaced by the current version, ''NeuroKit2''. A few major updates have been released since: * February 08, 2021: The 0.1.0 release coincides with the first publication of the software. * May 18, 2022: The 0.2.0 release coincides with an overhaul of the documentation. Features NeuroKit2 includes tools to work with cardiac activity from electrocardiography (ECG ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MATLAB
MATLAB (an abbreviation of "MATrix LABoratory") is a proprietary multi-paradigm programming language and numeric computing environment developed by MathWorks. MATLAB allows matrix manipulations, plotting of functions and data, implementation of algorithms, creation of user interfaces, and interfacing with programs written in other languages. Although MATLAB is intended primarily for numeric computing, an optional toolbox uses the MuPAD symbolic engine allowing access to symbolic computing abilities. An additional package, Simulink, adds graphical multi-domain simulation and model-based design for dynamic and embedded systems. As of 2020, MATLAB has more than 4 million users worldwide. They come from various backgrounds of engineering, science, and economics. History Origins MATLAB was invented by mathematician and computer programmer Cleve Moler. The idea for MATLAB was based on his 1960s PhD thesis. Moler became a math professor at the University of New Mexico and starte ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Magnetoencephalography
Magnetoencephalography (MEG) is a functional neuroimaging technique for mapping brain activity by recording magnetic fields produced by electrical currents occurring naturally in the brain, using very sensitive magnetometers. Arrays of SQUIDs (superconducting quantum interference devices) are currently the most common magnetometer, while the SERF (spin exchange relaxation-free) magnetometer is being investigated for future machines. Applications of MEG include basic research into perceptual and cognitive brain processes, localizing regions affected by pathology before surgical removal, determining the function of various parts of the brain, and neurofeedback. This can be applied in a clinical setting to find locations of abnormalities as well as in an experimental setting to simply measure brain activity. History MEG signals were first measured by University of Illinois physicist David Cohen in 1968, before the availability of the SQUID, using a copper induction coil as the d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Salk Institute
The Salk Institute for Biological Studies is a scientific research institute located in the La Jolla community of San Diego, California, U.S. The independent, non-profit institute was founded in 1960 by Jonas Salk, the developer of the polio vaccine; among the founding consultants were Jacob Bronowski and Francis Crick. Construction of the research facilities began in spring of 1962. The Salk Institute consistently ranks among the top institutions in the US in terms of research output and quality in the life sciences. In 2004, the ''Times Higher Education Supplement'' ranked Salk as the world's top biomedicine research institute, and in 2009 it was ranked number one globally by '' ScienceWatch'' in the neuroscience and behavior areas. The Salk Institute employs 850 researchers in 60 research groups and focuses its research in three areas: molecular biology and genetics; neurosciences; and plant biology. Research topics include aging, cancer, diabetes, birth defects, Alzheimer's dis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Graphical User Interface
The GUI ( "UI" by itself is still usually pronounced . or ), graphical user interface, is a form of user interface that allows users to interact with electronic devices through graphical icons and audio indicator such as primary notation, instead of text-based UIs, typed command labels or text navigation. GUIs were introduced in reaction to the perceived steep learning curve of CLIs ( command-line interfaces), which require commands to be typed on a computer keyboard. The actions in a GUI are usually performed through direct manipulation of the graphical elements. Beyond computers, GUIs are used in many handheld mobile devices such as MP3 players, portable media players, gaming devices, smartphones and smaller household, office and industrial controls. The term ''GUI'' tends not to be applied to other lower-display resolution types of interfaces, such as video games (where HUD (''head-up display'') is preferred), or not including flat screens like volumetric displays because ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Brain–computer Interface
A brain–computer interface (BCI), sometimes called a brain–machine interface (BMI) or smartbrain, is a direct communication pathway between the brain, brain's electrical activity and an external device, most commonly a computer or robotic limb. BCIs are often directed at researching, Brain mapping, mapping, assisting, Augmented cognition, augmenting, or repairing human Cognitive skill, cognitive or Sensory-motor coupling, sensory-motor functions. Implementations of BCIs range from non-invasive (Electroencephalography, EEG, Magnetoencephalography, MEG, Electrooculography, EOG, Magnetic resonance imaging, MRI) and partially invasive (Electrocorticography, ECoG and endovascular) to invasive (microelectrode array), based on how close electrodes get to brain tissue.Michael L Martini, BA, Eric Karl Oermann, MD, Nicholas L Opie, PhD, Fedor Panov, MD, Thomas Oxley, MD, PhD, Kurt Yaeger, MD, Sensor Modalities for Brain-Computer Interface Technology: A Comprehensive Literature Review, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |



.jpg)