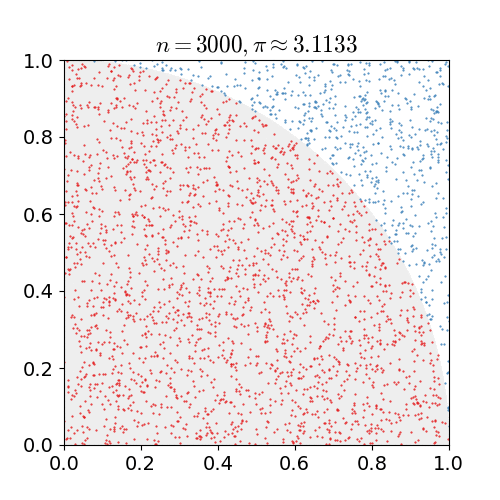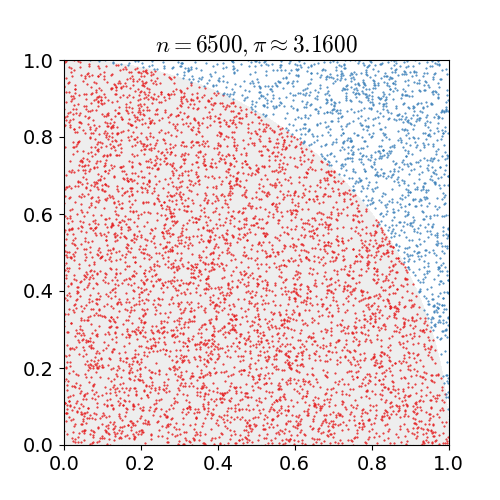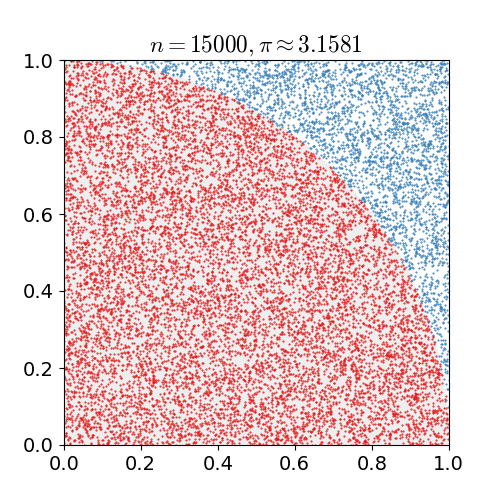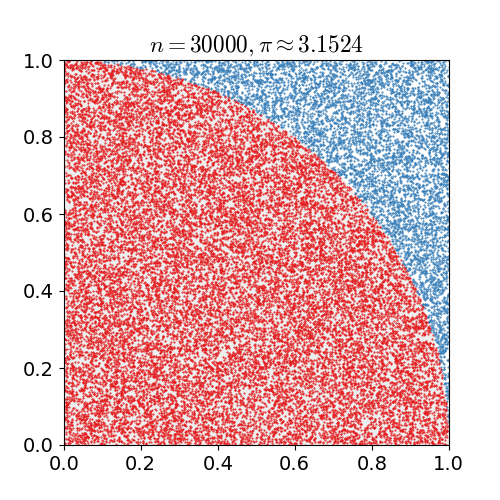
|
Monte Carlo Molecular Modeling
Monte Carlo molecular modelling is the application of Monte Carlo methods to molecular problems. These problems can also be modelled by the molecular dynamics method. The difference is that this approach relies on equilibrium statistical mechanics rather than molecular dynamics. Instead of trying to reproduce the dynamics of a system, it generates states according to appropriate Boltzmann distribution. Thus, it is the application of the Metropolis Monte Carlo simulation to molecular systems. It is therefore also a particular subset of the more general Monte Carlo method in statistical physics. It employs a Markov chain procedure in order to determine a new state for a system from a previous one. According to its stochastic nature, this new state is accepted at random. Each trial usually counts as a move. The avoidance of dynamics restricts the method to studies of static quantities only, but the freedom to choose moves makes the method very flexible. These moves must only satisfy a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Monte Carlo Method
Monte Carlo methods, or Monte Carlo experiments, are a broad class of computational algorithms that rely on repeated random sampling to obtain numerical results. The underlying concept is to use randomness to solve problems that might be deterministic in principle. They are often used in physical and mathematical problems and are most useful when it is difficult or impossible to use other approaches. Monte Carlo methods are mainly used in three problem classes: optimization, numerical integration, and generating draws from a probability distribution. In physics-related problems, Monte Carlo methods are useful for simulating systems with many coupled degrees of freedom, such as fluids, disordered materials, strongly coupled solids, and cellular structures (see cellular Potts model, interacting particle systems, McKean–Vlasov processes, kinetic models of gases). Other examples include modeling phenomena with significant uncertainty in inputs such as the calculation of ris ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
CP2K
CP2K is a freely available (GPL) quantum chemistry and solid state physics program package, written in Fortran 2008, to perform atomistic simulations of solid state, liquid, molecular, periodic, material, crystal, and biological systems. It provides a general framework for different methods: density functional theory (DFT) using a mixed Gaussian and plane waves approach (GPW) via LDA, GGA, MP2, or RPA levels of theory, classical pair and many-body potentials, semi-empirical ( AM1, PM3, MNDO, MNDOd, PM6) and tight-binding Hamiltonians, as well as Quantum Mechanics/Molecular Mechanics (QM/MM) hybrid schemes relying on the Gaussian Expansion of the Electrostatic Potential (GEEP). The Gaussian and Augmented Plane Waves method (GAPW) as an extension of the GPW method allows for all-electron calculations. CP2K can do simulations of molecular dynamics, metadynamics, Monte Carlo, Ehrenfest dynamics, vibrational analysis, core level spectroscopy, energy minimization, and transition sta ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Theoretical Chemistry
Theoretical chemistry is the branch of chemistry which develops theoretical generalizations that are part of the theoretical arsenal of modern chemistry: for example, the concepts of chemical bonding, chemical reaction, valence, the surface of potential energy, molecular orbitals, orbital interactions, and molecule activation. Overview Theoretical chemistry unites principles and concepts common to all branches of chemistry. Within the framework of theoretical chemistry, there is a systematization of chemical laws, principles and rules, their refinement and detailing, the construction of a hierarchy. The central place in theoretical chemistry is occupied by the doctrine of the interconnection of the structure and properties of molecular systems. It uses mathematical and physical methods to explain the structures and dynamics of chemical systems and to correlate, understand, and predict their thermodynamic and kinetic properties. In the most general sense, it is explanation of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Modelling
Molecular modelling encompasses all methods, theoretical and computational, used to model or mimic the behaviour of molecules. The methods are used in the fields of computational chemistry, drug design, computational biology and materials science to study molecular systems ranging from small chemical systems to large biological molecules and material assemblies. The simplest calculations can be performed by hand, but inevitably computers are required to perform molecular modelling of any reasonably sized system. The common feature of molecular modelling methods is the atomistic level description of the molecular systems. This may include treating atoms as the smallest individual unit (a molecular mechanics approach), or explicitly modelling protons and neutrons with its quarks, anti-quarks and gluons and electrons with its photons (a quantum chemistry approach). Molecular mechanics Molecular mechanics is one aspect of molecular modelling, as it involves the use of classical ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bond Fluctuation Model
The BFM (bond fluctuation model or bond fluctuation method) is a lattice model for simulating the conformation and dynamics of polymer systems. There are two versions of the BFM used: The earlier version was first introduced by I. Carmesin and Kurt Kremer in 1988, and the later version by J. Scott Shaffer in 1994. Conversion between models is possible. Model Carmesin and Kremer version In this model the monomers are represented by cubes on a regular cubic lattice with each cube occupying eight lattice positions. Each lattice position can only be occupied by one monomer in order to model excluded volume. The monomers are connected by a bond vector, which is taken from a set of typically 108 allowed vectors. There are different definitions for this vector set. One example for a bond vector set is made up from the six base vectors below using permutation and sign variation of the three vector components in each direction: : \mathbf = \mathbf \left( \begin 2 \\ 0 \\ 0 \end \right) \ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Software For Molecular Mechanics Modeling
This is a list of computer programs that are predominantly used for molecular mechanics calculations. See also *Car–Parrinello molecular dynamics *Comparison of force-field implementations *Comparison of nucleic acid simulation software *List of molecular graphics systems *List of protein structure prediction software *List of quantum chemistry and solid-state physics software *List of software for Monte Carlo molecular modeling *List of software for nanostructures modeling *Molecular design software *Molecular dynamics *Molecular modeling on GPUs *Molecule editor A molecule editor is a computer program for creating and modifying representations of chemical structures. Molecule editors can manipulate chemical structure representations in either a simulated two-dimensional space or three-dimensional space, v ... Notes and references External linksSINCRIS [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Software For Monte Carlo Molecular Modeling
This is a list of computer programs that use Monte Carlo methods for molecular modeling. * Abalone classical Hybrid MC * BOSS classical * Cassandra classical * CP2K * FEASST classical * GOMC classical * MacroModel classical * Materials Studio classical * ''ms''2classical *RASPA classical * QMCPACK quantum * Spartan classical * Tinker classical * Towhee classical{{cite journal, last=Martin, first=Marcus G., collaboration=Towhee, title= MCCCS Towhee: a tool for Monte Carlo molecular simulation., journal=Molecular Simulation, date=16 September 2013, volume=39, issue=14–15, pages=1212–1222, doi=10.1080/08927022.2013.828208, s2cid=97160184 See also * List of quantum chemistry and solid state physics software * Comparison of software for molecular mechanics modeling * Comparison of nucleic acid simulation software * Molecular design software * Molecule editor A molecule editor is a computer program for creating and modifying representations of chemical structures. Molecule edi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Quantum Monte Carlo
Quantum Monte Carlo encompasses a large family of computational methods whose common aim is the study of complex quantum systems. One of the major goals of these approaches is to provide a reliable solution (or an accurate approximation) of the quantum many-body problem. The diverse flavors of quantum Monte Carlo approaches all share the common use of the Monte Carlo method to handle the multi-dimensional integrals that arise in the different formulations of the many-body problem. Quantum Monte Carlo methods allow for a direct treatment and description of complex many-body effects encoded in the wave function, going beyond mean-field theory. In particular, there exist numerically exact and polynomially-scaling algorithms to exactly study static properties of boson systems without geometrical frustration. For fermions, there exist very good approximations to their static properties and numerically exact exponentially scaling quantum Monte Carlo algorithms, but none that are b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Simulated Annealing
Simulated annealing (SA) is a probabilistic technique for approximating the global optimum of a given function. Specifically, it is a metaheuristic to approximate global optimization in a large search space for an optimization problem. It is often used when the search space is discrete (for example the traveling salesman problem, the boolean satisfiability problem, protein structure prediction, and job-shop scheduling). For problems where finding an approximate global optimum is more important than finding a precise local optimum in a fixed amount of time, simulated annealing may be preferable to exact algorithms such as gradient descent or branch and bound. The name of the algorithm comes from annealing in metallurgy, a technique involving heating and controlled cooling of a material to alter its physical properties. Both are attributes of the material that depend on their thermodynamic free energy. Heating and cooling the material affects both the temperature and the the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Dynamics
Molecular dynamics (MD) is a computer simulation method for analyzing the physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a fixed period of time, giving a view of the dynamic "evolution" of the system. In the most common version, the trajectories of atoms and molecules are determined by numerically solving Newton's equations of motion for a system of interacting particles, where forces between the particles and their potential energies are often calculated using interatomic potentials or molecular mechanical force fields. The method is applied mostly in chemical physics, materials science, and biophysics. Because molecular systems typically consist of a vast number of particles, it is impossible to determine the properties of such complex systems analytically; MD simulation circumvents this problem by using numerical methods. However, long MD simulations are mathematically ill-conditioned, generating cumulative errors in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ising Model
The Ising model () (or Lenz-Ising model or Ising-Lenz model), named after the physicists Ernst Ising and Wilhelm Lenz, is a mathematical model of ferromagnetism in statistical mechanics. The model consists of discrete variables that represent magnetic dipole moments of atomic "spins" that can be in one of two states (+1 or −1). The spins are arranged in a graph, usually a lattice (where the local structure repeats periodically in all directions), allowing each spin to interact with its neighbors. Neighboring spins that agree have a lower energy than those that disagree; the system tends to the lowest energy but heat disturbs this tendency, thus creating the possibility of different structural phases. The model allows the identification of phase transitions as a simplified model of reality. The two-dimensional square-lattice Ising model is one of the simplest statistical models to show a phase transition. The Ising model was invented by the physicist , who gave it as a prob ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Detailed Balance
The principle of detailed balance can be used in kinetic systems which are decomposed into elementary processes (collisions, or steps, or elementary reactions). It states that at equilibrium, each elementary process is in equilibrium with its reverse process. History The principle of detailed balance was explicitly introduced for collisions by Ludwig Boltzmann. In 1872, he proved his H-theorem using this principle.Boltzmann, L. (1964), Lectures on gas theory, Berkeley, CA, USA: U. of California Press. The arguments in favor of this property are founded upon microscopic reversibility. Tolman, R. C. (1938). ''The Principles of Statistical Mechanics''. Oxford University Press, London, UK. Five years before Boltzmann, James Clerk Maxwell used the principle of detailed balance for gas kinetics with the reference to the principle of sufficient reason. He compared the idea of detailed balance with other types of balancing (like cyclic balance) and found that "Now it is impossible to as ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |