|
List Of Software For Monte Carlo Molecular Modeling
This is a list of computer programs that use Monte Carlo methods for molecular modeling. * Abalone classical Hybrid MC * BOSS classical * Cassandra classical * CP2K * FEASST classical * GOMC classical * MacroModel classical * Materials Studio classical * ''ms''2classical *RASPA classical * QMCPACK quantum * Spartan classical * Tinker classical * Towhee classical{{cite journal, last=Martin, first=Marcus G., collaboration=Towhee, title= MCCCS Towhee: a tool for Monte Carlo molecular simulation., journal=Molecular Simulation, date=16 September 2013, volume=39, issue=14–15, pages=1212–1222, doi=10.1080/08927022.2013.828208, s2cid=97160184 See also * List of quantum chemistry and solid state physics software * Comparison of software for molecular mechanics modeling * Comparison of nucleic acid simulation software * Molecular design software * Molecule editor A molecule editor is a computer program for creating and modifying representations of chemical structures. Molecule edi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Monte Carlo Method
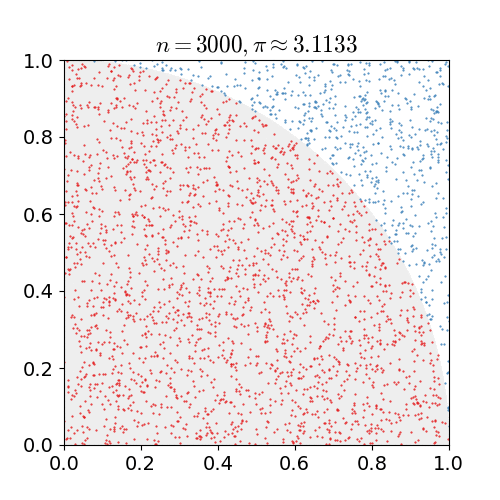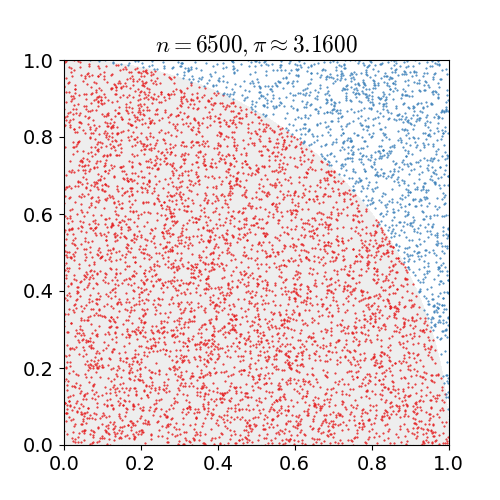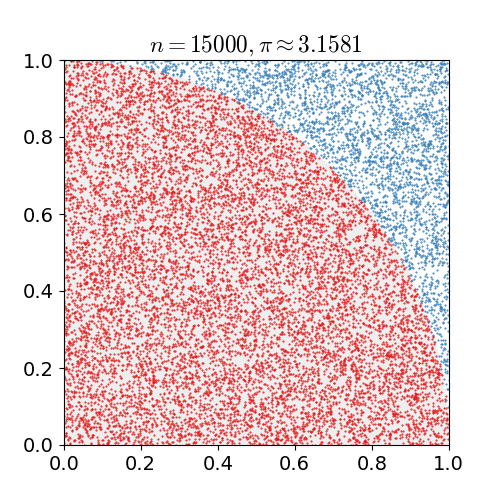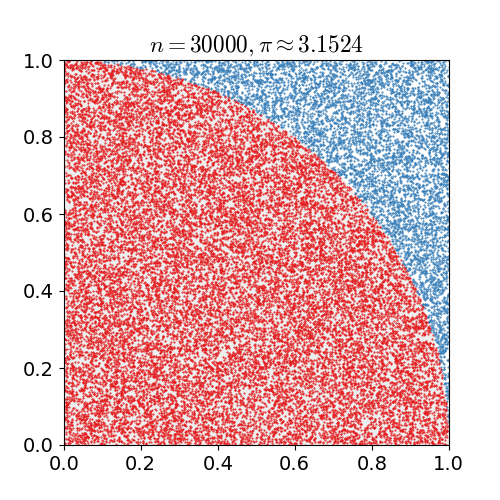
Monte Carlo methods, or Monte Carlo experiments, are a broad class of computational algorithms that rely on repeated random sampling to obtain numerical results. The underlying concept is to use randomness to solve problems that might be deterministic in principle. They are often used in physical and mathematical problems and are most useful when it is difficult or impossible to use other approaches. Monte Carlo methods are mainly used in three problem classes: optimization, numerical integration, and generating draws from a probability distribution. In physics-related problems, Monte Carlo methods are useful for simulating systems with many coupled degrees of freedom, such as fluids, disordered materials, strongly coupled solids, and cellular structures (see cellular Potts model, interacting particle systems, McKean–Vlasov processes, kinetic models of gases). Other examples include modeling phenomena with significant uncertainty in inputs such as the calculation of ris ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Spartan (chemistry Software)
Spartan is a molecular modelling and computational chemistry application from Wavefunction. It contains code for molecular mechanics, semi-empirical methods, ''ab initio'' models, density functional models, post-Hartree–Fock models, and thermochemical recipes including G3(MP2) and T1. Quantum chemistry calculations in Spartan are powered by Q-Chem. Primary functions are to supply information about structures, relative stabilities and other properties of isolated molecules. Molecular mechanics calculations on complex molecules are common in the chemical community. Quantum chemical calculations, including Hartree–Fock method molecular orbital calculations, but especially calculations that include electronic correlation, are more time-consuming in comparison. Quantum chemical calculations are also called upon to furnish information about mechanisms and product distributions of chemical reactions, either directly by calculations on transition states, or based on Hammond's p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecule Editor
A molecule editor is a computer program for creating and modifying representations of chemical structures. Molecule editors can manipulate chemical structure representations in either a simulated two-dimensional space or three-dimensional space, via 2D computer graphics or 3D computer graphics, respectively. Two-dimensional output is used as illustrations or to query chemical databases. Three-dimensional output is used to build molecular models, usually as part of molecular modelling software packages. Database molecular editors such as Leatherface, RECAP, and Molecule Slicer allow large numbers of molecules to be modified automatically according to rules such as 'deprotonate carboxylic acids' or 'break exocyclic bonds' that can be specified by a user. Molecule editors typically support reading and writing at least one file format or line notation. Examples of each include Molfile and simplified molecular input line entry specification (SMILES), respectively. Files generated by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Design Software
Molecular design software is notable software for molecular modeling, that provides special support for developing molecular models ''de novo''. In contrast to the usual molecular modeling programs, such as for molecular dynamics and quantum chemistry, such software ''directly'' supports the aspects related to constructing molecular models, including: * Molecular graphics * interactive molecular drawing and conformational editing * building polymeric molecules, crystals, and solvated systems * partial charges development * geometry optimization * support for the different aspects of force field development Comparison of software covering the major aspects of molecular design Notes and references See also {{columns-list, colwidth=30em, *Molecule editor *Molecular modelling *Molecular modeling on GPUs *Protein design *Drug design *Force field (chemistry) *Comparison of force field implementations *Comparison of nucleic acid simulation software *Comparison of software for ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Comparison Of Nucleic Acid Simulation Software
This is a list of notable computer programs that are used for nucleic acid Nucleic acids are biopolymers, macromolecules, essential to all known forms of life. They are composed of nucleotides, which are the monomers made of three components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main cl ...s simulations. See also References Computational chemistry software Software comparisons Molecular dynamics software Molecular modelling software {{science-software-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Comparison Of Software For Molecular Mechanics Modeling
This is a list of computer programs that are predominantly used for molecular mechanics calculations. See also *Car–Parrinello molecular dynamics *Comparison of force-field implementations *Comparison of nucleic acid simulation software *List of molecular graphics systems *List of protein structure prediction software *List of quantum chemistry and solid-state physics software * List of software for Monte Carlo molecular modeling * List of software for nanostructures modeling *Molecular design software *Molecular dynamics *Molecular modeling on GPUs *Molecule editor A molecule editor is a computer program for creating and modifying representations of chemical structures. Molecule editors can manipulate chemical structure representations in either a simulated two-dimensional space or three-dimensional space, v ... Notes and references External linksSINCRIS [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
List Of Quantum Chemistry And Solid State Physics Software
Quantum chemistry computer programs are used in computational chemistry to implement the methods of quantum chemistry. Most include the Hartree–Fock (HF) and some post-Hartree–Fock methods. They may also include density functional theory (DFT), molecular mechanics or semi-empirical quantum chemistry methods. The programs include both open source Open source is source code that is made freely available for possible modification and redistribution. Products include permission to use the source code, design documents, or content of the product. The open-source model is a decentralized sof ... and commercial software. Most of them are large, often containing several separate programs, and have been developed over many years. Overview The following tables illustrates some of the main capabilities of notable packages: Numerical details Quantum chemistry and solid-state physics characteristics Post processing packages in quantum chemistry and solid-state physics ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tinker (software)
Tinker, previously stylized as TINKER, is a suite of computer software applications for molecular dynamics simulation. The codes provide a complete and general set of tools for molecular mechanics and molecular dynamics, with some special features for biomolecules. The core of the software is a modular set of callable routines which allow manipulating coordinates and evaluating potential energy and derivatives via straightforward means. Tinker works on Windows, macOS, Linux and Unix. The source code is available free of charge to non-commercial users under a proprietary license. The code is written in portable FORTRAN 77, Fortran 95 or CUDA with common extensions, and some C. Core developers are: (athe Jay Ponder lab at the Department of Chemistry, Washington University in St. Louis, St. Louis, Missouri. Laboratory head Ponder is Full Professor of Chemistry, and of Biochemistry & Molecular Biophysics; (bthe Pengyu Ren lab at the Department of Biomedical Engineering Universit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ms2 (software)
''ms''2 is a non-commercial molecular simulation program. It comprises both molecular dynamics and Monte Carlo simulation algorithms. ''ms''2 is designed for the calculation of thermodynamic properties of fluids. A large number of thermodynamic properties can be readily computed using ''ms''2, e.g. phase equilibrium, transport and caloric properties. ''ms''2 is limited to homogeneous state simulations. Features ''ms''2 contains two molecular simulation techniques: molecular dynamics (MD) and Monte-Carlo. ''ms''2 supports the calculation of vapor-liquid equilibria of pure components as well as multi-component mixtures. Different Phase equilibrium calculation methods are implemented in ''ms''2. Furthermore, ''ms''2 is capable of sampling various classical ensembles such as NpT, NVE, NVT, NpH. To evaluate the chemical potential, Widom's test molecule method and thermodynamic integration are implemented. Also, algorithms for the sampling of transport properties are implemented in ' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Modeling
Molecular modelling encompasses all methods, theoretical and computational, used to model or mimic the behaviour of molecules. The methods are used in the fields of computational chemistry, drug design, computational biology and materials science to study molecular systems ranging from small chemical systems to large biological molecules and material assemblies. The simplest calculations can be performed by hand, but inevitably computers are required to perform molecular modelling of any reasonably sized system. The common feature of molecular modelling methods is the atomistic level description of the molecular systems. This may include treating atoms as the smallest individual unit (a molecular mechanics approach), or explicitly modelling protons and neutrons with its quarks, anti-quarks and gluons and electrons with its photons (a quantum chemistry approach). Molecular mechanics Molecular mechanics is one aspect of molecular modelling, as it involves the use of classical mec ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Materials Studio
Materials Studio is software for simulating and modeling materials. It is developed and distributed by BIOVIA (formerly Accelrys), a firm specializing in research software for computational chemistry, bioinformatics, cheminformatics, molecular dynamics simulation, and quantum mechanics. This software is used in advanced research of various materials, such as polymers, carbon nanotubes, catalysts, metals, ceramics, and so on, by universities (e.g., North Dakota State University), research centers, and high tech companies. Materials Studio is a client–server model software package with Microsoft Windows-based PC clients and Windows and Linux-based servers running on PCs, Linux IA-64 workstations (including Silicon Graphics (SGI) Altix) and HP XC clusters. Software components *Analytical and Crystallization: to investigate, predict, and modify crystal structure and crystal growth. **Morphology **Polymorph Predictor **Reflex, Reflex Plus, Reflex QPA: to assist the interpretat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MacroModel
MacroModel is a computer program for molecular modelling of organic compounds and biopolymers. It features various chemistry force fields, plus energy minimizing algorithms, to predict geometry and relative conformational energies of molecules. MacroModel is maintained by Schrödinger, LLC. It performs simulations in the framework of classical mechanics, also termed ''molecular mechanics'', and can perform molecular dynamics simulations to model systems at finite temperatures using stochastic dynamics and mixed Monte Carlo algorithms. ''MacroModel'' supports Windows, Linux, macOS, Silicon Graphics (SGI) IRIX, and IBM AIX AIX (Advanced Interactive eXecutive, pronounced , "ay-eye-ex") is a series of proprietary Unix operating systems developed and sold by IBM for several of its computer platforms. Background Originally released for the IBM RT PC RISC .... The Macromodel software package was first been described in the scientific literature in 1990, and has b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |