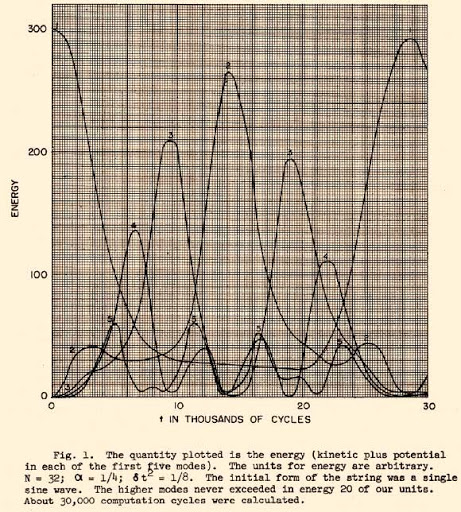
|
MacroModel
MacroModel is a computer program for molecular modelling of organic compounds and biopolymers. It features various chemistry force fields, plus energy minimizing algorithms, to predict geometry and relative conformational energies of molecules. MacroModel is maintained by Schrödinger, LLC. It performs simulations in the framework of classical mechanics, also termed ''molecular mechanics'', and can perform molecular dynamics simulations to model systems at finite temperatures using stochastic dynamics and mixed Monte Carlo algorithms. ''MacroModel'' supports Windows, Linux, macOS, Silicon Graphics (SGI) IRIX, and IBM AIX AIX (Advanced Interactive eXecutive, pronounced , "ay-eye-ex") is a series of proprietary Unix operating systems developed and sold by IBM for several of its computer platforms. Background Originally released for the IBM RT PC RISC .... The Macromodel software package was first been described in the scientific literature in 1990, and has b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Mechanics
Molecular mechanics uses classical mechanics to model molecular systems. The Born–Oppenheimer approximation is assumed valid and the potential energy of all systems is calculated as a function of the nuclear coordinates using force fields. Molecular mechanics can be used to study molecule systems ranging in size and complexity from small to large biological systems or material assemblies with many thousands to millions of atoms. All-atomistic molecular mechanics methods have the following properties: * Each atom is simulated as one particle * Each particle is assigned a radius (typically the van der Waals radius), polarizability, and a constant net charge (generally derived from quantum calculations and/or experiment) * Bonded interactions are treated as ''springs'' with an equilibrium distance equal to the experimental or calculated bond length Variants on this theme are possible. For example, many simulations have historically used a ''united-atom'' representation in which ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Free Energy Perturbation
Free energy perturbation (FEP) is a method based on statistical mechanics that is used in computational chemistry for computing free energy differences from molecular dynamics or Metropolis Monte Carlo simulations. The FEP method was introduced by Robert W. Zwanzig in 1954. According to the free-energy perturbation method, the free energy difference for going from state A to state B is obtained from the following equation, known as the ''Zwanzig equation'': :\Delta F(\mathbf \rightarrow \mathbf) = F_\mathbf - F_\mathbf = -k_\mathrm T \ln \left \langle \exp \left ( - \frac \right ) \right \rangle _\mathbf where ''T'' is the temperature, ''k''B is Boltzmann's constant, and the angular brackets denote an average over a simulation run for state A. In practice, one runs a normal simulation for state A, but each time a new configuration is accepted, the energy for state B is also computed. The difference between states A and B may be in the atom types involved, in which case the Δ''F ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Dynamics
Molecular dynamics (MD) is a computer simulation method for analyzing the physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a fixed period of time, giving a view of the dynamic "evolution" of the system. In the most common version, the trajectories of atoms and molecules are determined by numerically solving Newton's equations of motion for a system of interacting particles, where forces between the particles and their potential energies are often calculated using interatomic potentials or molecular mechanical force fields. The method is applied mostly in chemical physics, materials science, and biophysics. Because molecular systems typically consist of a vast number of particles, it is impossible to determine the properties of such complex systems analytically; MD simulation circumvents this problem by using numerical methods. However, long MD simulations are mathematically ill-conditioned, generating cumulative errors in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Comparison Of Nucleic Acid Simulation Software
This is a list of notable computer programs that are used for nucleic acid Nucleic acids are biopolymers, macromolecules, essential to all known forms of life. They are composed of nucleotides, which are the monomers made of three components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main cl ...s simulations. See also References Computational chemistry software Software comparisons Molecular dynamics software Molecular modelling software {{science-software-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Dynamics
Molecular dynamics (MD) is a computer simulation method for analyzing the physical movements of atoms and molecules. The atoms and molecules are allowed to interact for a fixed period of time, giving a view of the dynamic "evolution" of the system. In the most common version, the trajectories of atoms and molecules are determined by numerically solving Newton's equations of motion for a system of interacting particles, where forces between the particles and their potential energies are often calculated using interatomic potentials or molecular mechanical force fields. The method is applied mostly in chemical physics, materials science, and biophysics. Because molecular systems typically consist of a vast number of particles, it is impossible to determine the properties of such complex systems analytically; MD simulation circumvents this problem by using numerical methods. However, long MD simulations are mathematically ill-conditioned, generating cumulative errors in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
OPLS
The OPLS (Optimized Potentials for Liquid Simulations) force field was developed by Prof. William L. Jorgensen at Purdue University and later at Yale University. Functional form The functional form of the OPLS force field is very similar to that of AMBER: E \left(r^N \right ) = E_\mathrm + E_\mathrm + E_\mathrm + E_\mathrm E_\mathrm = \sum_\mathrm K_r (r-r_0)^2 \, E_\mathrm = \sum_\mathrm k_\theta (\theta-\theta_0)^2 \, E_\mathrm = \sum_\mathrm \left( \frac \left 1 + \cos (\phi-\phi_1) \right + \frac \left 1 - \cos (2\phi-\phi_2) \right + \frac \left 1 + \cos (3\phi-\phi_3) \right + \frac \left 1 - \cos (4\phi-\phi_4) \right \right) E_\mathrm = \sum_ f_ \left( \frac - \frac + \frac \right) with the combining rules A_ = \sqrt and C_ = \sqrt. Intramolecular nonbonded interactions E_\mathrm are counted only for atoms three or more bonds apart; 1,4 interactions are sca ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
AMBER
Amber is fossilized tree resin that has been appreciated for its color and natural beauty since Neolithic times. Much valued from antiquity to the present as a gemstone, amber is made into a variety of decorative objects."Amber" (2004). In Maxine N. Lurie and Marc Mappen (eds.) ''Encyclopedia of New Jersey'', Rutgers University Press, . Amber is used in jewelry and has been used as a healing agent in folk medicine. There are five classes of amber, defined on the basis of their chemical constituents. Because it originates as a soft, sticky tree resin, amber sometimes contains animal and plant material as inclusions. Amber occurring in coal seams is also called resinite, and the term ''ambrite'' is applied to that found specifically within New Zealand coal seams. Etymology The English word ''amber'' derives from Arabic (ultimately from Middle Persian ''ambar'') via Middle Latin ''ambar'' and Middle French ''ambre''. The word was adopted in Middle English in the 14th century ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Force Field (chemistry)
In the context of chemistry and molecular modelling, a force field is a computational method that is used to estimate the forces between atoms within molecules and also between molecules. More precisely, the force field refers to the functional form and parameter sets used to calculate the potential energy of a system of atoms or coarse-grained particles in molecular mechanics, molecular dynamics, or Monte Carlo simulations. The parameters for a chosen energy function may be derived from experiments in physics and chemistry, calculations in quantum mechanics, or both. Force fields are interatomic potentials and utilize the same concept as force fields in classical physics, with the difference that the force field parameters in chemistry describe the energy landscape, from which the acting forces on every particle are derived as a gradient of the potential energy with respect to the particle coordinates. ''All-atom'' force fields provide parameters for every type of atom in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Implicit Solvation
Implicit solvation (sometimes termed continuum solvation) is a method to represent solvent as a continuous medium instead of individual “explicit” solvent molecules, most often used in molecular dynamics simulations and in other applications of molecular mechanics. The method is often applied to estimate free energy of solute-solvent interactions in structural and chemical processes, such as folding or conformational transitions of proteins, DNA, RNA, and polysaccharides, association of biological macromolecules with ligands, or transport of drugs across biological membranes. The implicit solvation model is justified in liquids, where the potential of mean force can be applied to approximate the averaged behavior of many highly dynamic solvent molecules. However, the interfaces and the interiors of biological membranes or proteins can also be considered as media with specific solvation or dielectric properties. These media are not necessarily uniform, since their properties ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
IBM AIX
AIX (Advanced Interactive eXecutive, pronounced , "ay-eye-ex") is a series of Proprietary software, proprietary Unix operating systems developed and sold by IBM for several of its computer platforms. Background Originally released for the IBM RT PC RISC workstation in 1986, AIX has supported a wide variety of hardware platforms, including the IBM RS/6000 series and later IBM Power microprocessors, Power and PowerPC-based systems, IBM System i, System/370 mainframes, IBM Personal System/2, PS/2 personal computers, and the Apple Network Server. It is currently supported on IBM Power Systems alongside IBM i and Linux. AIX is based on UNIX System V with 4.3BSD-compatible extensions. It is certified to the UNIX 03 and UNIX V7 marks of the Single UNIX Specification, beginning with AIX versions 5.3 and 7.2 TL5 respectively. Older versions were previously certified to the UNIX 95 and UNIX 98 marks. AIX was the first operating system to have a journaling file system, and IBM has c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
IRIX
IRIX ( ) is a discontinued operating system developed by Silicon Graphics (SGI) to run on the company's proprietary MIPS workstations and servers. It is based on UNIX System V with BSD extensions. In IRIX, SGI originated the XFS file system and the industry-standard OpenGL graphics system. History SGI originated the IRIX name in the 1988 release 3.0 of the operating system for the SGI IRIS 4D series of workstations and servers. Previous releases are identified only by the release number prefixed by "4D1-", such as "4D1-2.2". The "4D1-" prefix continued to be used in official documentation to prefix IRIX release numbers. Prior to the IRIS 4D, SGI bundled the GL2 operating system, based on UniSoft UniPlus System V Unix, and using the proprietary MEX (Multiple EXposure) windowing system. IRIX 3.x is based on UNIX System V Release 3 with 4.3BSD enhancements, and incorporates the 4Sight windowing system, based on NeWS and IRIS GL. SGI's own Extent File System (EFS) replaces the S ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |