|
H3K9ac
H3K9ac is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the acetylation at the 9th lysine residue of the histone H3 protein. The H3K9 histone has two jobs. Genes get turned on if this mark is acetylated and silences them if methylated. H3K9ac is an important acetylation and connected with active promoters. H3K9ac and H3K14ac have been shown to be part of the active promoter state. They are also present over bivalent promoters and active enhancers. This is also a mark for liver cancer through a defect in the H3K9ac/H3K9me3 transition. Lysine acetylation and deacetylation Proteins are typically acetylated on lysine residues and this reaction relies on acetyl-coenzyme A as the acetyl group donor. In histone acetylation and deacetylation, histone proteins are acetylated and deacetylated on lysine residues in the N-terminal tail as part of gene regulation. Typically, these reactions are catalyzed by enzymes with ''histone ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
H3K14ac
H3K14ac is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the acetylation at the 14th lysine residue of the histone H3 protein. H3K14ac has not been widely studied partly due to previous lack of commercially available antibody. H3K9ac and H3K14ac have been shown to be part of the active promoter state. They are also present over bivalent promoters and active enhancers. H3K14ac is also enriched over a subset of inactive promoters. The Tudor domain of the H3K9 methyltransferase SETDB1 binds to methylated H3 with both K14 acetylation and K9 methylation. SETDB1 silences retroviruses and gene regulation. Lysine acetylation and deacetylation Proteins are typically acetylated on lysine residues and this reaction relies on acetyl-coenzyme A as the acetyl group donor. In histone acetylation and deacetylation, histone proteins are acetylated and deacetylated on lysine residues in the N-terminal tail as part of gene regulation. Typ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
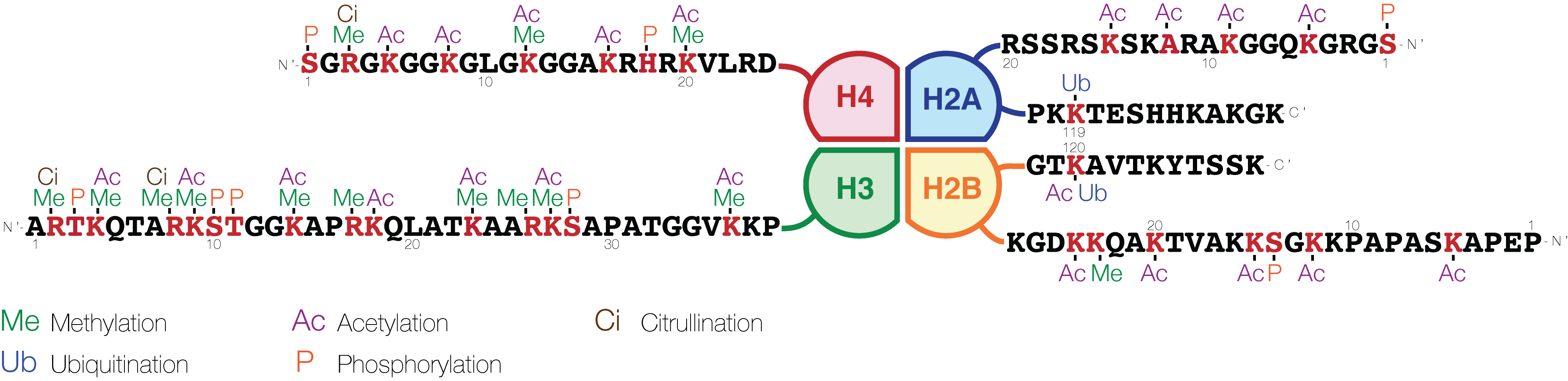
Histone Code
The histone code is a hypothesis that the transcription of genetic information encoded in DNA is in part regulated by chemical modifications (known as ''histone marks'') to histone proteins, primarily on their unstructured ends. Together with similar modifications such as DNA methylation it is part of the epigenetic code. Histones associate with DNA to form nucleosomes, which themselves bundle to form chromatin fibers, which in turn make up the more familiar chromosome. Histones are globular proteins with a flexible N-terminus (taken to be the tail) that protrudes from the nucleosome. Many of the histone tail modifications correlate very well to chromatin structure and both histone modification state and chromatin structure correlate well to gene expression levels. The critical concept of the histone code hypothesis is that the histone modifications serve to recruit other proteins by specific recognition of the modified histone via protein domains specialized for such purposes ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone Acetylation And Deacetylation
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are wrapped into 30- nanometer fibers that form tightly packed chromatin. Histones prevent DNA from becoming tangled and protect it from DNA damage. In addition, histones play important roles in gene regulation and DNA replication. Without histones, unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA if completely stretched out; however, when wound about histones, this length is reduced to about 90 micrometers (0.09 mm) of 30 nm diameter chromatin fibers. There are five families of histones which are designated H1/H5 (linker histones), H2, H3, and H4 (core histones). The nucleosome core is formed of two H2A-H2B dimers and a H3-H4 tetramer. The tight wrapping of DNA around hist ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histones
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are wrapped into 30-nanometer fibers that form tightly packed chromatin. Histones prevent DNA from becoming tangled and protect it from DNA damage. In addition, histones play important roles in gene regulation and DNA replication. Without histones, unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA if completely stretched out; however, when wound about histones, this length is reduced to about 90 micrometers (0.09 mm) of 30 nm diameter chromatin fibers. There are five families of histones which are designated H1/H5 (linker histones), H2, H3, and H4 (core histones). The nucleosome core is formed of two H2A-H2B dimers and a H3-H4 tetramer. The tight wrapping of DNA around histones ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epigenetic
In biology, epigenetics is the study of stable phenotypic changes (known as ''marks'') that do not involve alterations in the DNA sequence. The Greek prefix '' epi-'' ( "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in addition to" the traditional genetic basis for inheritance. Epigenetics most often involves changes that affect the regulation of gene expression, but the term can also be used to describe any heritable phenotypic change. Such effects on cellular and physiological phenotypic traits may result from external or environmental factors, or be part of normal development. The term also refers to the mechanism of changes: functionally relevant alterations to the genome that do not involve mutation of the nucleotide sequence. Examples of mechanisms that produce such changes are DNA methylation and histone modification, each of which alters how genes are expressed without altering the underlying DNA sequence. Gene expression can ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methylation
In the chemical sciences, methylation denotes the addition of a methyl group on a substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These terms are commonly used in chemistry, biochemistry, soil science, and the biological sciences. In biological systems, methylation is catalyzed by enzymes; such methylation can be involved in modification of heavy metals, regulation of gene expression, regulation of protein function, and RNA processing. In vitro methylation of tissue samples is also one method for reducing certain histological staining artifacts. The reverse of methylation is demethylation. In biology In biological systems, methylation is accomplished by enzymes. Methylation can modify heavy metals, regulate gene expression, RNA processing and protein function. It has been recognized as a key process underlying epigenetics. Methanogenesis Methanogenesis, the process th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ENCODE
The Encyclopedia of DNA Elements (ENCODE) is a public research project which aims to identify functional elements in the human genome. ENCODE also supports further biomedical research by "generating community resources of genomics data, software, tools and methods for genomics data analysis, and products resulting from data analyses and interpretations." The current phase of ENCODE (2016-2019) is adding depth to its resources by growing the number of cell types, data types, assays and now includes support for examination of the mouse genome. History ENCODE was launched by the US National Human Genome Research Institute (NHGRI) in September 2003. Intended as a follow-up to the Human Genome Project, the ENCODE project aims to identify all functional elements in the human genome. The project involves a worldwide consortium of research groups, and data generated from this project can be accessed through public databases. The initial release of ENCODE was in 2013 and since has be ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
H3K36me3
H3K36me3 is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the tri- methylation at the 36th lysine residue of the histone H3 protein and often associated with gene bodies. There are diverse modifications at H3K36 and have many important biological processes. H3K36 has different acetylation and methylation states with no similarity to each other. Nomenclature H3K36me3 indicates trimethylation of lysine 36 on histone H3 protein subunit: Lysine Methylation This diagram shows the progressive methylation of a lysine residue. The tri-methylation denotes the methylation present in H3K36me3. Understanding histone modifications The genomic DNA of eukaryotic cells is wrapped around special protein molecules known as Histones. The complexes formed by the looping of the DNA are known as chromatin. The basic structural unit of chromatin is the nucleosome: this consists of the core octamer of histones (H2A, H2B, H3 and H4) as well ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone proteins and resembles thread wrapped around a spool. The nucleosome is the fundamental subunit of chromatin. Each nucleosome is composed of a little less than two turns of DNA wrapped around a set of eight proteins called histones, which are known as a histone octamer. Each histone octamer is composed of two copies each of the histone proteins H2A, H2B, H3, and H4. DNA must be compacted into nucleosomes to fit within the cell nucleus. In addition to nucleosome wrapping, eukaryotic chromatin is further compacted by being folded into a series of more complex structures, eventually forming a chromosome. Each human cell contains about 30 million nucleosomes. Nucleosomes are thought to carry epigenetically inherited information in the form of covalent modifications of their core histones. Nucleosome positions in the gen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromatin
Chromatin is a complex of DNA and protein found in eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important roles in reinforcing the DNA during cell division, preventing DNA damage, and regulating gene expression and DNA replication. During mitosis and meiosis, chromatin facilitates proper segregation of the chromosomes in anaphase; the characteristic shapes of chromosomes visible during this stage are the result of DNA being coiled into highly condensed chromatin. The primary protein components of chromatin are histones. An octamer of two sets of four histone cores (Histone H2A, Histone H2B, Histone H3, and Histone H4) bind to DNA and function as "anchors" around which the strands are wound.Maeshima, K., Ide, S., & Babokhov, M. (2019). Dynamic chromatin organization without the 30-nm fiber. ''Current opinion in cell biology, 58,'' 95–104. https://doi.o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Acetyl Group
In organic chemistry, acetyl is a functional group with the chemical formula and the structure . It is sometimes represented by the symbol Ac (not to be confused with the element actinium). In IUPAC nomenclature, acetyl is called ethanoyl, although this term is barely heard. The acetyl group contains a methyl group () single-bonded to a carbonyl (). The carbonyl center of an acyl radical has one nonbonded electron with which it forms a chemical bond to the remainder ''R'' of the molecule. The acetyl moiety is a component of many organic compounds, including acetic acid, the neurotransmitter acetylcholine, acetyl-CoA, acetylcysteine, acetaminophen (also known as paracetamol), and acetylsalicylic acid (also known as aspirin). Acetylation In nature The introduction of an acetyl group into a molecule is called acetylation. In biological organisms, acetyl groups are commonly transferred from acetyl-CoA to other organic molecules. Acetyl-CoA is an intermediate both ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amino Acid Residue
Protein structure is the molecular geometry, three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, the monomers of the polymer. A single amino acid monomer may also be called a ''residue'' indicating a repeating unit of a polymer. Proteins form by amino acids undergoing condensation reactions, in which the amino acids lose one water molecule per chemical reaction, reaction in order to attach to one another with a peptide bond. By convention, a chain under 30 amino acids is often identified as a peptide, rather than a protein. To be able to perform their biological function, proteins fold into one or more specific spatial conformations driven by a number of non-covalent interactions such as hydrogen bonding, ionic interactions, Van der Waals forces, and hydrophobic packing. To understand the functions of proteins at a molecular level, it is often necessary to determine their Pro ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |