|
DHRS7B
Dehydrogenase/reductase (SDR family) member 7B is an enzyme encoded by the DHRS7B gene in humans, found on chromosome 17p11.2. DHRS7B encodes a protein that is predicted to function in steroid hormone regulation. A deletion in the chromosomal region 17p11.2 has been associated with Smith-Magenis Syndrome, a genetic developmental disorder. Gene Overview The DHRS7B gene is located on the positive strand of chromosome 17, beginning at position 21030258 and ending at position 21094836 (64579 bp). DHRS7B contains seven exons with no predicted alternate splice forms, resulting in an 1841 bp mRNA product. Upstream of DHRS7B on the negative strand of chromosome 17p11.2 are the genes ''Coiled-coil domain containing 144 family, N-terminal-like'' ( CCDC144NL) and ''Ubiquitin specific peptidase 22'' (USP22). Downstream of DHSRS7B on the negative strand of chromosome 17p11.2 is the gene ''Transmembrane protein 11'' ( TMEM11), and on the positive strand is the gene ''Mitogen-activated pr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Enzyme
Enzymes () are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called ''enzymology'' and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties. Enzymes are known to catalyze more than 5,000 biochemical reaction types. Other biocatalysts are catalytic RNA molecules, called ribozymes. Enzymes' specificity comes from their unique three-dimensional structures. Like all catalysts, enzymes increase the reaction ra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
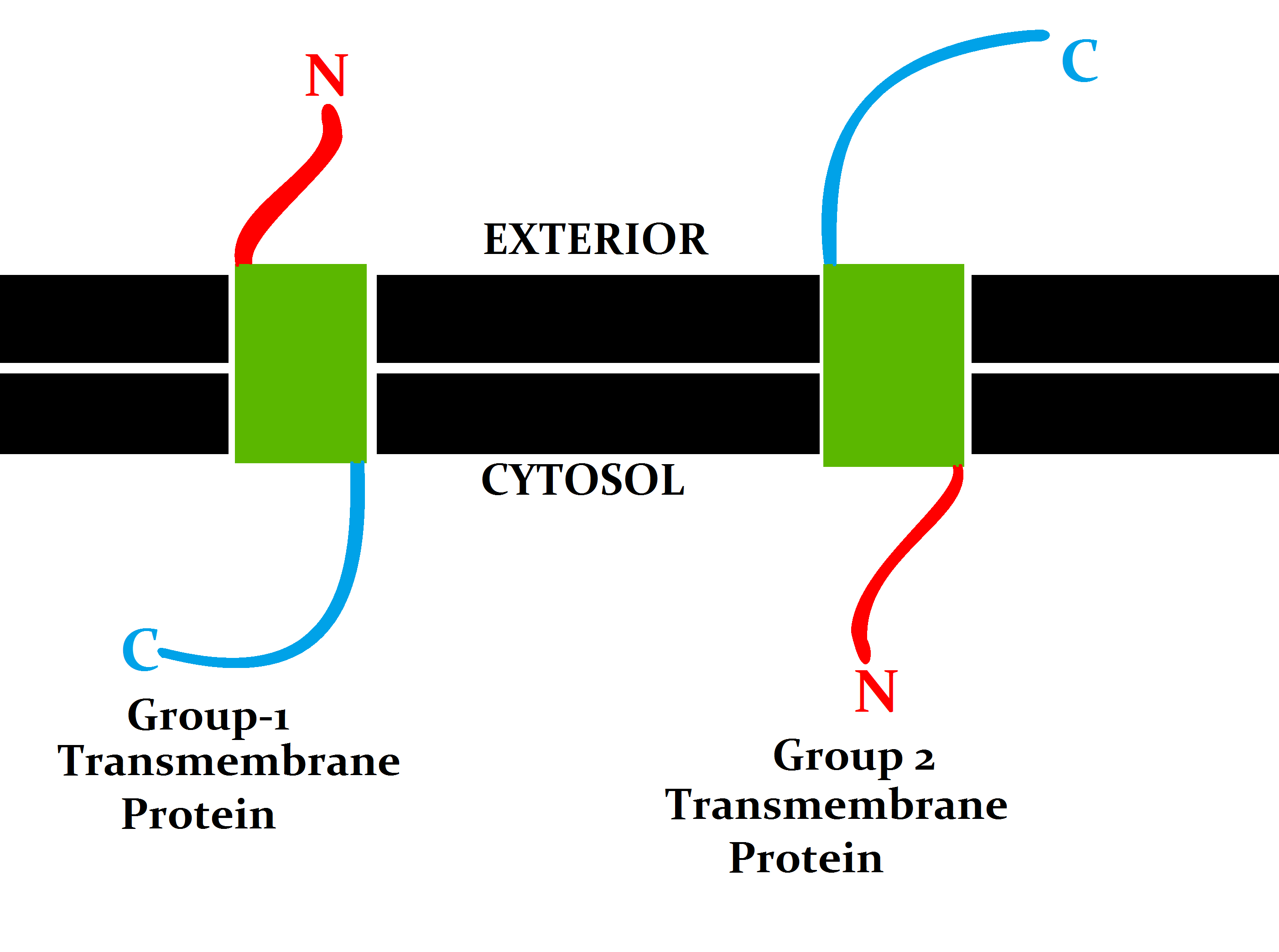
Transmembrane
A transmembrane protein (TP) is a type of integral membrane protein that spans the entirety of the cell membrane. Many transmembrane proteins function as gateways to permit the transport of specific substances across the membrane. They frequently undergo significant conformational changes to move a substance through the membrane. They are usually highly hydrophobic and aggregate and precipitate in water. They require detergents or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents. The peptide sequence that spans the membrane, or the transmembrane segment, is largely hydrophobic and can be visualized using the hydropathy plot. Depending on the number of transmembrane segments, transmembrane proteins can be classified as single-span (or bitopic) or multi-span (polytopic). Some other integral membrane proteins are called monotopic, meaning that they are also permanently attached to the membrane, but do not pass ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reptiles
Reptiles, as most commonly defined are the animals in the Class (biology), class Reptilia ( ), a paraphyletic grouping comprising all sauropsid, sauropsids except birds. Living reptiles comprise turtles, crocodilians, Squamata, squamates (lizards and snakes) and rhynchocephalians (tuatara). As of March 2022, the Reptile Database includes about 11,700 species. In the traditional Linnaean taxonomy, Linnaean classification system, birds are considered a separate class to reptiles. However, crocodilians are more closely related to birds than they are to other living reptiles, and so modern Cladistics, cladistic classification systems include birds within Reptilia, redefining the term as a clade. Other cladistic definitions abandon the term reptile altogether in favor of the clade Sauropsida, which refers to all amniotes more closely related to modern reptiles than to mammals. The study of the traditional reptile Order (biology), orders, historically combined with that of modern amphi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mammals
Mammals () are a group of vertebrate animals constituting the class Mammalia (), characterized by the presence of mammary glands which in females produce milk for feeding (nursing) their young, a neocortex (a region of the brain), fur or hair, and three middle ear bones. These characteristics distinguish them from reptiles (including birds) from which they diverged in the Carboniferous, over 300 million years ago. Around 6,400 extant species of mammals have been described divided into 29 orders. The largest orders, in terms of number of species, are the rodents, bats, and Eulipotyphla (hedgehogs, moles, shrews, and others). The next three are the Primates (including humans, apes, monkeys, and others), the Artiodactyla ( cetaceans and even-toed ungulates), and the Carnivora (cats, dogs, seals, and others). In terms of cladistics, which reflects evolutionary history, mammals are the only living members of the Synapsida (synapsids); this clade, together with Saur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
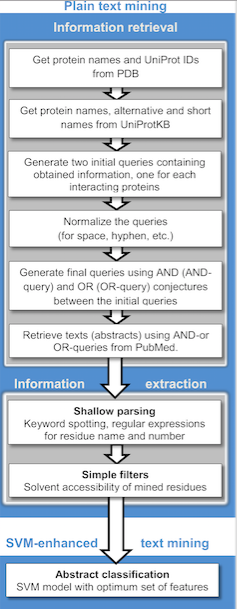
Text Mining
Text mining, also referred to as ''text data mining'', similar to text analytics, is the process of deriving high-quality information from text. It involves "the discovery by computer of new, previously unknown information, by automatically extracting information from different written resources." Written resources may include websites, books, emails, reviews, and articles. High-quality information is typically obtained by devising patterns and trends by means such as statistical pattern learning. According to Hotho et al. (2005) we can distinguish between three different perspectives of text mining: information extraction, data mining, and a KDD (Knowledge Discovery in Databases) process. Text mining usually involves the process of structuring the input text (usually parsing, along with the addition of some derived linguistic features and the removal of others, and subsequent insertion into a database), deriving patterns within the structured data, and finally evaluation and inte ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Coimmunoprecipitation
Immunoprecipitation (IP) is the technique of precipitating a protein antigen out of solution using an antibody that specifically binds to that particular protein. This process can be used to isolate and concentrate a particular protein from a sample containing many thousands of different proteins. Immunoprecipitation requires that the antibody be coupled to a solid substrate at some point in the procedure. Types Individual protein immunoprecipitation (IP) Involves using an antibody that is specific for a known protein to isolate that particular protein out of a solution containing many different proteins. These solutions will often be in the form of a crude lysate of a plant or animal tissue. Other sample types could be body fluids or other samples of biological origin. Protein complex immunoprecipitation (Co-IP) Immunoprecipitation of intact protein complexes (i.e. antigen along with any proteins or ligands that are bound to it) is known as co-immunoprecipitation (Co-I ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histones
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are wrapped into 30-nanometer fibers that form tightly packed chromatin. Histones prevent DNA from becoming tangled and protect it from DNA damage. In addition, histones play important roles in gene regulation and DNA replication. Without histones, unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA if completely stretched out; however, when wound about histones, this length is reduced to about 90 micrometers (0.09 mm) of 30 nm diameter chromatin fibers. There are five families of histones which are designated H1/H5 (linker histones), H2, H3, and H4 (core histones). The nucleosome core is formed of two H2A-H2B dimers and a H3-H4 tetramer. The tight wrapping of DNA around histones ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitinated
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A. The addition of ubiquitin to a substrate protein is called ubiquitylation (or, alternatively, ubiquitination or ubiquitinylation). Ubiquitylation affects proteins in many ways: it can mark them for degradation via the proteasome, alter their cellular location, affect their activity, and promote or prevent protein interactions. Ubiquitylation involves three main steps: activation, conjugation, and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), respectively. The result of this sequential cascade is to bind ubiquitin to lysine residues on the protein substrate via an isopeptide bond, c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that synthesizes RNA from a DNA template. Using the enzyme helicase, RNAP locally opens the double-stranded DNA so that one strand of the exposed nucleotides can be used as a template for the synthesis of RNA, a process called transcription. A transcription factor and its associated transcription mediator complex must be attached to a DNA binding site called a promoter region before RNAP can initiate the DNA unwinding at that position. RNAP not only initiates RNA transcription, it also guides the nucleotides into position, facilitates attachment and elongation, has intrinsic proofreading and replacement capabilities, and termination recognition capability. In eukaryotes, RNAP can build chains as long as 2.4 million nucleotides. RNAP produces RNA that, functionally, is either for protein coding, i.e. messenger RNA (mRNA); or n ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Two Hybrid Screening
Two-hybrid screening (originally known as yeast two-hybrid system or Y2H) is a molecular biology technique used to discover protein–protein interactions (PPIs) and protein–DNA interactions by testing for physical interactions (such as binding) between two proteins or a single protein and a DNA molecule, respectively. The premise behind the test is the activation of downstream reporter gene(s) by the binding of a transcription factor onto an upstream activating sequence (UAS). For two-hybrid screening, the transcription factor is split into two separate fragments, called the DNA-binding domain (DBD or often also abbreviated as BD) and activating domain (AD). The BD is the domain responsible for binding to the UAS and the AD is the domain responsible for the activation of transcription. The Y2H is thus a protein-fragment complementation assay. History Pioneered by Stanley Fields and Ok-Kyu Song in 1989, the technique was originally designed to detect protein–protein i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BRE (gene)
BRCA1-A complex subunit BRE is a protein that in humans is encoded by the ''BRE'' gene. Repair of DNA damage BRE, the protein product of the BRE (gene), is a core component of the deubiquitin complex BRCA1-A. Other core components of the BRCA1-A complex are the BRCC36 protein (BRCC3 gene), MERIT40 protein (BABAM1 gene), and RAP80 protein ( UIMC1 gene). BRCA1, as distinct from BRCA1-A, is employed in the repair of chromosomal damage with an important role in the error-free homologous recombinational (HR) repair of DNA double-strand breaks. Sequestration of BRCA1 away from the DNA damage site suppresses homologous recombination and redirects the cell in the direction of repair by the process of non-homologous end joining (NHEJ). The role of BRCA1-A appears to be to bind BRCA1 with high affinity and withdraw it away from the site of DNA damage to the periphery where it remains sequestered, thus promoting NHEJ in preference to HR. Protein-protein interactions BRE (gene) has b ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MED19
Mediator complex subunit 19 (Med19) is a protein that in humans is encoded by the MED19 gene. Function The protein encoded by this gene is a subunit of the Mediator complex, which binds to gene-specific regulatory factors and provides support for the basal RNA polymerase II transcription machinery. This gene has been implicated in the growth of several types of cancer, and inhibition of its expression inhibits the growth and spread of these cancers. Two transcript variants encoding different isoforms have been found for this gene. rovided by RefSeq, Nov 2015 See also *Mediator Mediator may refer to: *A person who engages in mediation *Business mediator, a mediator in business * Vanishing mediator, a philosophical concept * Mediator variable, in statistics Chemistry and biology *Mediator (coactivator), a multiprotein ... References Further reading * * * * * * * * * {{NLM content ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |