|
Coimmunoprecipitation
Immunoprecipitation (IP) is the technique of precipitating a protein antigen out of solution using an antibody that specifically binds to that particular protein. This process can be used to isolate and concentrate a particular protein from a sample containing many thousands of different proteins. Immunoprecipitation requires that the antibody be coupled to a solid substrate at some point in the procedure. Types Individual protein immunoprecipitation (IP) Involves using an antibody that is specific for a known protein to isolate that particular protein out of a solution containing many different proteins. These solutions will often be in the form of a crude lysate of a plant or animal tissue. Other sample types could be body fluids or other samples of biological origin. Protein complex immunoprecipitation (Co-IP) Immunoprecipitation of intact protein complexes (i.e. antigen along with any proteins or ligands that are bound to it) is known as co-immunoprecipitation (Co-I ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Precipitation (chemistry)
In an aqueous solution, precipitation is the process of transforming a dissolved chemical substance, substance into an insoluble solid from a Supersaturated solution, super-saturated solution. The solid formed is called the precipitate. In case of an inorganic chemical reaction leading to precipitation, the chemical reagent causing the solid to form is called the ''precipitant''. The clear liquid remaining above the precipitated or the centrifuged solid phase is also called the 'supernate' or 'supernatant'. The notion of precipitation can also be extended to other domains of chemistry (organic chemistry and biochemistry) and even be applied to the solid phases (''e.g.'', metallurgy and alloys) when solid impurities Segregation (materials science), segregate from a solid phase. Supersaturation The precipitation of a compound may occur when its concentration exceeds its solubility. This can be due to temperature changes, solvent evaporation, or by mixing solvents. Precipitatio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cross-link
In chemistry and biology a cross-link is a bond or a short sequence of bonds that links one polymer chain to another. These links may take the form of covalent bonds or ionic bonds and the polymers can be either synthetic polymers or natural polymers (such as proteins). In polymer chemistry "cross-linking" usually refers to the use of cross-links to promote a change in the polymers' physical properties. When "crosslinking" is used in the biological field, it refers to the use of a probe to link proteins together to check for protein–protein interactions, as well as other creative cross-linking methodologies. Although the term is used to refer to the "linking of polymer chains" for both sciences, the extent of crosslinking and specificities of the crosslinking agents vary greatly. As with all science, there are overlaps, and the following delineations are a starting point to understanding the subtleties. Polymer chemistry Crosslinking is the general term for the process of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PAR-CLIP
PAR-CLIP (photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation) is a biochemistry, biochemical method for identifying the binding sites of cellular RNA-binding proteins (RBPs) and microRNA-containing ribonucleoprotein complexes (miRNPs). The method relies on the incorporation of ribonucleoside analogs that are photosensitivity, photoreactive, such as 4-thiouridine (4-SU) and 6-thioguanosine (6-SG), into nascent RNA transcripts by living cells. Irradiation of the cells by ultraviolet light of 365 nm wavelength induces efficient cross-link, crosslinking of photoreactive nucleoside–flow tracer, labeled cellular RNAs to interacting RBPs. Immunoprecipitation of the RBP of interest is followed by isolation of the crosslinked and coimmunoprecipitated RNA. The isolated RNA is converted into a complementary DNA, cDNA library and is deep sequencing, deep sequenced using next-generation sequencing technology. Recently, PAR-CLIP have been applied to determine the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RT-PCR
Reverse transcription polymerase chain reaction (RT-PCR) is a laboratory technique combining reverse transcription of RNA into DNA (in this context called complementary DNA or cDNA) and amplification of specific DNA targets using polymerase chain reaction (PCR). It is primarily used to measure the amount of a specific RNA. This is achieved by monitoring the amplification reaction using fluorescence, a technique called real-time PCR or quantitative PCR (qPCR). Combined RT-PCR and qPCR are routinely used for analysis of gene expression and quantification of viral RNA in research and clinical settings. The close association between RT-PCR and qPCR has led to metonymic use of the term qPCR to mean RT-PCR. Such use may be confusing, as RT-PCR can be used without qPCR, for example to enable molecular cloning, sequencing or simple detection of RNA. Conversely, qPCR may be used without RT-PCR, for example to quantify the copy number of a specific piece of DNA. Nomenclature The combine ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery. Knowledge of DNA sequences has become indispensable for basic biological research, DNA Genographic Projects and in numerous applied fields such as medical diagnosis, biotechnology, forensic biology, virology and biological systematics. Comparing healthy and mutated DNA sequences can diagnose different diseases including various cancers, characterize antibody repertoire, and can be used to guide patient treatment. Having a quick way to sequence DNA allows for faster and more individualized medical care to be administered, and for more organisms to be identified and cataloged. The rapid speed of sequencing attained with modern D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
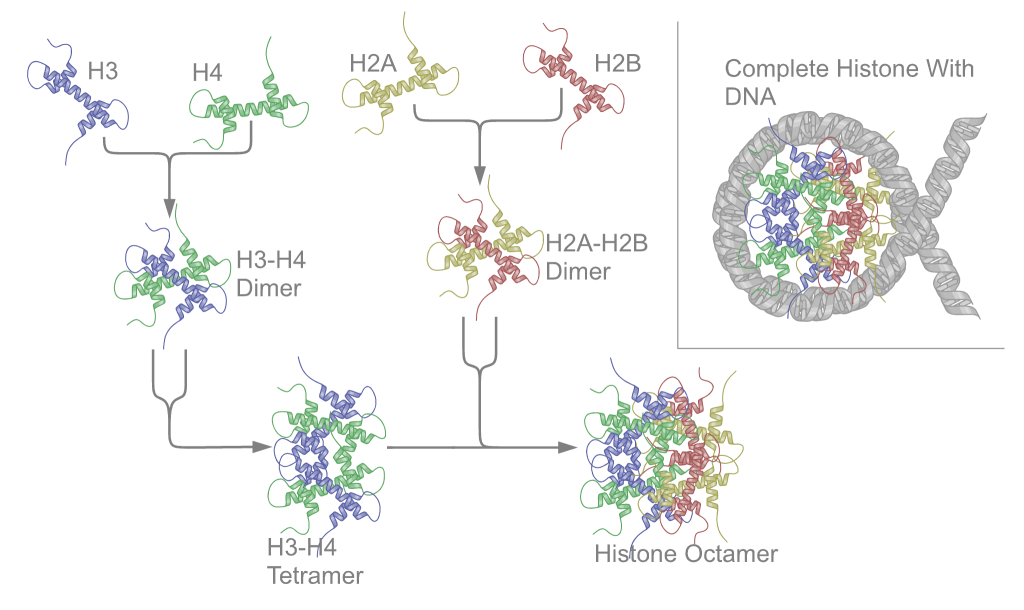
Ribonucleoproteins
Nucleoproteins are proteins conjugated with nucleic acids (either DNA or RNA). Typical nucleoproteins include ribosomes, nucleosomes and viral nucleocapsid proteins. Structures Nucleoproteins tend to be positively charged, facilitating interaction with the negatively charged nucleic acid chains. The tertiary structures and biological functions of many nucleoproteins are understood.Graeme K. Hunter G. K. (2000): Vital Forces. The discovery of the molecular basis of life. Academic Press, London 2000, . Important techniques for determining the structures of nucleoproteins include X-ray diffraction, nuclear magnetic resonance and cryo-electron microscopy. Viruses Virus genomes (either DNA or RNA) are extremely tightly packed into the viral capsid. Many viruses are therefore little more than an organised collection of nucleoproteins with their binding sites pointing inwards. Structurally characterised viral nucleoproteins include influenza, rabies, Ebola, Bunyamwera, Schmallen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ChIP-on-chip
ChIP-on-chip (also known as ChIP-chip) is a technology that combines chromatin immunoprecipitation ('ChIP') with DNA microarray (''"chip"''). Like regular ChIP, ChIP-on-chip is used to investigate interactions between proteins and DNA ''in vivo''. Specifically, it allows the identification of the cistrome, the sum of binding sites, for DNA-binding proteins on a genome-wide basis. Whole-genome analysis can be performed to determine the locations of binding sites for almost any protein of interest. As the name of the technique suggests, such proteins are generally those operating in the context of chromatin. The most prominent representatives of this class are transcription factors, replication-related proteins, like origin recognition complex protein (ORC), histones, their variants, and histone modifications. The goal of ChIP-on-chip is to locate protein binding sites that may help identify functional elements in the genome. For example, in the case of a transcription factor as a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Microarray
A DNA microarray (also commonly known as DNA chip or biochip) is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome. Each DNA spot contains picomoles (10−12 moles) of a specific DNA sequence, known as '' probes'' (or ''reporters'' or '' oligos''). These can be a short section of a gene or other DNA element that are used to hybridize a cDNA or cRNA (also called anti-sense RNA) sample (called ''target'') under high-stringency conditions. Probe-target hybridization is usually detected and quantified by detection of fluorophore-, silver-, or chemiluminescence-labeled targets to determine relative abundance of nucleic acid sequences in the target. The original nucleic acid arrays were macro arrays approximately 9 cm × 12 cm and the first computerized image based analysis was published in 1981. It was inv ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cistrome
In simple words, the cistrome refers a collection of regulatory elements of a set of genes, including transcription factor binding-sites and histone modifications. More specifically, "the set of cis-acting targets of a trans-acting factor on a genome-wide scale, also known as the ''in vivo'' genome-wide location of transcription factor binding-sites or histone modifications". The term cistrome is a portmanteau of cistr (from cistron) + ome (from genome). The term cistrome was coined by investigators at the Dana–Farber Cancer Institute and Harvard Medical School. Technologies such as chromatin immunoprecipitation combined with microarray analysis "ChIP-on-chip" or with massively parallel DNA sequencing "ChIP-Seq" have greatly facilitated the definition of the cistrome of transcription factors and other chromatin Chromatin is a complex of DNA and protein found in eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chip-Sequencing
ChIP-sequencing, also known as ChIP-seq, is a method used to analyze protein interactions with DNA. ChIP-seq combines chromatin immunoprecipitation (ChIP) with massively parallel DNA sequencing to identify the binding sites of DNA-associated proteins. It can be used to map global binding sites precisely for any protein of interest. Previously, ChIP-on-chip was the most common technique utilized to study these protein–DNA relations. Uses ChIP-seq is primarily used to determine how transcription factors and other chromatin-associated proteins influence phenotype-affecting mechanisms. Determining how proteins interact with DNA to regulate gene expression is essential for fully understanding many biological processes and disease states. This epigenetic information is complementary to genotype and expression analysis. ChIP-seq technology is currently seen primarily as an alternative to ChIP-chip which requires a hybridization array. This introduces some bias, as an array is restrict ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Plasmid
A plasmid is a small, extrachromosomal DNA molecule within a cell that is physically separated from chromosomal DNA and can replicate independently. They are most commonly found as small circular, double-stranded DNA molecules in bacteria; however, plasmids are sometimes present in archaea and eukaryotic organisms. In nature, plasmids often carry genes that benefit the survival of the organism and confer selective advantage such as antibiotic resistance. While chromosomes are large and contain all the essential genetic information for living under normal conditions, plasmids are usually very small and contain only additional genes that may be useful in certain situations or conditions. Artificial plasmids are widely used as vectors in molecular cloning, serving to drive the replication of recombinant DNA sequences within host organisms. In the laboratory, plasmids may be introduced into a cell via transformation. Synthetic plasmids are available for procurement over the inter ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is a method widely used to rapidly make millions to billions of copies (complete or partial) of a specific DNA sample, allowing scientists to take a very small sample of DNA and amplify it (or a part of it) to a large enough amount to study in detail. PCR was invented in 1983 by the American biochemist Kary Mullis at Cetus Corporation; Mullis and biochemist Michael Smith (chemist), Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993. PCR is fundamental to many of the procedures used in genetic testing and research, including analysis of Ancient DNA, ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory research for a broad variety of applications ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |