ChIP-on-chip on:
[Wikipedia]
[Google]
[Amazon]
 ChIP-on-chip (also known as ChIP-chip) is a technology that combines
ChIP-on-chip (also known as ChIP-chip) is a technology that combines
 In the first step, the protein of interest (POI) is
In the first step, the protein of interest (POI) is
 After a sufficiently large time frame to allow hybridization, the array is illuminated with fluorescent light. Those probes on the array that are hybridized to one of the labeled fragments emit a light signal that is captured by a camera. This image contains all raw data for the remaining part of the workflow.
This raw data, encoded as false-color image, needs to be converted to numerical values before the actual analysis can be done. The analysis and information extraction of the raw data often remains the most challenging part for ChIP-on-chip experiments. Problems arise throughout this portion of the workflow, ranging from the initial chip read-out, to suitable methods to subtract background noise, and finally to appropriate
After a sufficiently large time frame to allow hybridization, the array is illuminated with fluorescent light. Those probes on the array that are hybridized to one of the labeled fragments emit a light signal that is captured by a camera. This image contains all raw data for the remaining part of the workflow.
This raw data, encoded as false-color image, needs to be converted to numerical values before the actual analysis can be done. The analysis and information extraction of the raw data often remains the most challenging part for ChIP-on-chip experiments. Problems arise throughout this portion of the workflow, ranging from the initial chip read-out, to suitable methods to subtract background noise, and finally to appropriate
Chip-on-Chip (CoC)
Package Information from Amkor Technology
CoCAS: a free Analysis software for Agilent ChIP-on-Chip experiments
rMAT: R implementation from MAT program to normalize and analyze tiling arrays and ChIP-chip data. {{DEFAULTSORT:Chip-On-Chip Genomics techniques Molecular biology Molecular biology techniques Protein methods Bioinformatics Microarrays
chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) is a type of immunoprecipitation experimental technique used to investigate the interaction between proteins and DNA in the cell. It aims to determine whether specific proteins are associated with specific genom ...
('ChIP') with DNA microarray
A DNA microarray (also commonly known as a DNA chip or biochip) is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or t ...
(''"chip"''). Like regular ChIP, ChIP-on-chip is used to investigate interactions between protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
s and DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
''in vivo
Studies that are ''in vivo'' (Latin for "within the living"; often not italicized in English) are those in which the effects of various biological entities are tested on whole, living organisms or cells, usually animals, including humans, an ...
''. Specifically, it allows the identification of the cistrome, the sum of binding site
In biochemistry and molecular biology, a binding site is a region on a macromolecule such as a protein that binds to another molecule with specificity. The binding partner of the macromolecule is often referred to as a ligand. Ligands may includ ...
s, for DNA-binding proteins on a genome-wide basis. Whole-genome analysis can be performed to determine the locations of binding sites for almost any protein of interest. As the name of the technique suggests, such proteins are generally those operating in the context of chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important r ...
. The most prominent representatives of this class are transcription factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription (genetics), transcription of genetics, genetic information from DNA to messenger RNA, by binding t ...
s, replication-related proteins, like origin recognition complex
In molecular biology, origin recognition complex (ORC) is a multi-subunit DNA binding complex (6 subunits) that binds in all eukaryotes and archaea in an Adenosine triphosphate, ATP-dependent manner to origins of replication. The subunits of this ...
protein (ORC), histone
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei and in most Archaeal phyla. They act as spools around which DNA winds to create structural units called nucleosomes ...
s, their variants, and histone modifications.
The goal of ChIP-on-chip is to locate protein binding sites that may help identify functional elements in the genome
A genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding genes, other functional regions of the genome such as ...
. For example, in the case of a transcription factor as a protein of interest, one can determine its transcription factor binding sites throughout the genome. Other proteins allow the identification of promoter regions, enhancers
In genetics, an enhancer is a short (50–1500 bp) region of DNA that can be bound by proteins ( activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcriptio ...
, repressors and silencing elements, insulators
Insulator may refer to:
* Insulator (electricity), a substance that resists electricity
** Pin insulator, a device that isolates a wire from a physical support such as a pin on a utility pole
** Strain insulator, a device that is designed to work ...
, boundary elements, and sequences that control DNA replication. If histones are subject of interest, it is believed that the distribution of modifications and their localizations may offer new insights into the mechanisms of regulation
Regulation is the management of complex systems according to a set of rules and trends. In systems theory, these types of rules exist in various fields of biology and society, but the term has slightly different meanings according to context. Fo ...
.
One of the long-term goals ChIP-on-chip was designed for is to establish a catalogue of (selected) organisms that lists all protein-DNA interactions under various physiological conditions. This knowledge would ultimately help in the understanding of the machinery behind gene regulation, cell proliferation
Cell proliferation is the process by which ''a cell grows and divides to produce two daughter cells''. Cell proliferation leads to an exponential increase in cell number and is therefore a rapid mechanism of tissue growth. Cell proliferation ...
, and disease progression. Hence, ChIP-on-chip offers both potential to complement our knowledge about the orchestration of the genome on the nucleotide level and information on higher levels of information and regulation as it is propagated by research on epigenetics
In biology, epigenetics is the study of changes in gene expression that happen without changes to the DNA sequence. The Greek prefix ''epi-'' (ἐπι- "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in ...
.
__TOC__
Technological platforms
The technical platforms to conduct ChIP-on-chip experiments areDNA microarray
A DNA microarray (also commonly known as a DNA chip or biochip) is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or t ...
s, or ''"chips"''. They can be classified and distinguished according to various characteristics:
''Probe type'': DNA arrays can comprise either mechanically spotted cDNA
In genetics, complementary DNA (cDNA) is DNA that was reverse transcribed (via reverse transcriptase) from an RNA (e.g., messenger RNA or microRNA). cDNA exists in both single-stranded and double-stranded forms and in both natural and engin ...
s or PCR-products, mechanically spotted oligonucleotide
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, Recombinant DNA, research, and Forensic DNA, forensics. Commonly made in the laboratory by Oligonucleotide synthesis, solid-phase ...
s, or oligonucleotides that are synthesized ''in situ
is a Latin phrase meaning 'in place' or 'on site', derived from ' ('in') and ' ( ablative of ''situs'', ). The term typically refers to the examination or occurrence of a process within its original context, without relocation. The term is use ...
''. The early versions of microarrays were designed to detect RNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA and deoxyrib ...
s from expressed genomic regions (open reading frame
In molecular biology, reading frames are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a prokaryotic DNA sequence, where only one of the six possible reading frames ...
s aka ORFs). Although such arrays are perfectly suited to study gene expression profiles, they have limited importance in ChIP experiments since most "interesting" proteins with respect to this technique bind in intergenic region
An intergenic region is a stretch of DNA sequences located between genes. Intergenic regions may contain functional elements and junk DNA.
Properties and functions
Intergenic regions may contain a number of functional DNA sequences such as p ...
s. Nowadays, even custom-made arrays can be designed and fine-tuned to match the requirements of an experiment. Also, any sequence of nucleotides can be synthesized to cover genic as well as intergenic regions.
''Probe size'': Early version of cDNA arrays had a probe length of about 200bp. Latest array versions use oligos as short as 70- (Microarrays, Inc.) to 25-mers (Affymetrix
Affymetrix is now Applied Biosystems, a brand of DNA microarray products sold by Thermo Fisher Scientific that originated with an American biotechnology research and development and manufacturing company of the same name. The Santa Clara, Calif ...
). (Feb 2007)
''Probe composition'': There are tiled and non-tiled DNA arrays. Non-tiled arrays use probes selected according to non-spatial criteria, i.e., the DNA sequences used as probes have no fixed distances in the genome. Tiled arrays, however, select a genomic region (or even a whole genome) and divide it into equal chunks. Such a region is called tiled path. The average distance between each pair of neighboring chunks (measured from the center of each chunk) gives the resolution of the tiled path. A path can be overlapping, end-to-end or spaced.
''Array size'': The first microarrays used for ChIP-on-Chip contained about 13,000 spotted DNA segments representing all ORFs and intergenic regions from the yeast genome. Nowadays, Affymetrix offers whole-genome tiled yeast arrays with a resolution of 5bp (all in all 3.2 million probes). Tiled arrays for the human genome become more and more powerful, too. Just to name one example, Affymetrix offers a set of seven arrays with about 90 million probes, spanning the complete non-repetitive part of the human genome with about 35bp spacing. (Feb 2007)
Besides the actual microarray, other hard- and software equipment is necessary to run ChIP-on-chip experiments. It is generally the case that one company's microarrays can not be analyzed by another company's processing hardware. Hence, buying an array requires also buying the associated workflow equipment. The most important elements are, among others, hybridization ovens, chip scanners, and software packages for subsequent numerical analysis of the raw data.
Workflow of a ChIP-on-chip experiment
Starting with a biological question, a ChIP-on-chip experiment can be divided into three major steps: The first is to set up and design the experiment by selecting the appropriate array and probe type. Second, the actual experiment is performed in the wet-lab. Last, during the dry-lab portion of the cycle, gathered data are analyzed to either answer the initial question or lead to new questions so that the cycle can start again.Wet-lab portion of the workflow
 In the first step, the protein of interest (POI) is
In the first step, the protein of interest (POI) is cross-link
In chemistry and biology, a cross-link is a bond or a short sequence of bonds that links one polymer chain to another. These links may take the form of covalent bonds or ionic bonds and the polymers can be either synthetic polymers or natural ...
ed with the DNA site it binds to in an ''in vitro
''In vitro'' (meaning ''in glass'', or ''in the glass'') Research, studies are performed with Cell (biology), cells or biological molecules outside their normal biological context. Colloquially called "test-tube experiments", these studies in ...
'' environment. Usually this is done by a gentle formaldehyde
Formaldehyde ( , ) (systematic name methanal) is an organic compound with the chemical formula and structure , more precisely . The compound is a pungent, colourless gas that polymerises spontaneously into paraformaldehyde. It is stored as ...
fixation that is reversible with heat.
Then, the cells are lysed and the DNA is sheared by sonication
image:Sonicator.jpg, A sonicator at the Weizmann Institute of Science during sonicationSonication is the act of applying sound energy to agitate particles in a sample, for various purposes such as the extraction of multiple compounds from plants, ...
or using micrococcal nuclease. This results in double-stranded chunks of DNA fragments, normally 1 kb or less in length. Those that were cross-link
In chemistry and biology, a cross-link is a bond or a short sequence of bonds that links one polymer chain to another. These links may take the form of covalent bonds or ionic bonds and the polymers can be either synthetic polymers or natural ...
ed to the POI form a POI-DNA complex.
In the next step, only these complexes are filtered out of the set of DNA fragments, using an antibody
An antibody (Ab) or immunoglobulin (Ig) is a large, Y-shaped protein belonging to the immunoglobulin superfamily which is used by the immune system to identify and neutralize antigens such as pathogenic bacteria, bacteria and viruses, includin ...
specific to the POI. The antibodies may be attached to a solid surface, may have a magnetic bead, or some other physical property that allows separation of cross-linked complexes and unbound fragments. This procedure is essentially an immunoprecipitation
Immunoprecipitation (IP) is the technique of precipitating a protein antigen out of solution using an antibody that specifically binds to that particular protein. This process can be used to isolate and concentrate a particular protein from a sam ...
(IP) of the protein. This can be done either by using a tagged protein with an antibody against the tag (ex. FLAG
A flag is a piece of textile, fabric (most often rectangular) with distinctive colours and design. It is used as a symbol, a signalling device, or for decoration. The term ''flag'' is also used to refer to the graphic design employed, and fla ...
, HA, c-myc) or with an antibody to the native protein.
The cross-linking of POI-DNA complexes is reversed (usually by heating) and the DNA strands are purified. For the rest of the workflow, the POI is no longer necessary.
After an amplification and denaturation step, the single-stranded DNA fragments are labeled with a fluorescent
Fluorescence is one of two kinds of photoluminescence, the emission of light by a substance that has absorbed light or other electromagnetic radiation. When exposed to ultraviolet radiation, many substances will glow (fluoresce) with color ...
tag such as Cy5 or Alexa 647.
Finally, the fragments are poured over the surface of the DNA microarray, which is spotted with short, single-stranded sequences that cover the genomic portion of interest. Whenever a labeled fragment "finds" a complementary fragment on the array, they will hybridize and form again a double-stranded DNA fragment.
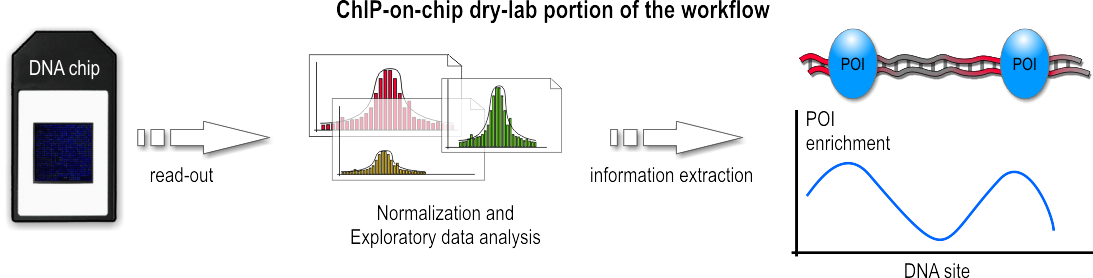
Dry-lab portion of the workflow
 After a sufficiently large time frame to allow hybridization, the array is illuminated with fluorescent light. Those probes on the array that are hybridized to one of the labeled fragments emit a light signal that is captured by a camera. This image contains all raw data for the remaining part of the workflow.
This raw data, encoded as false-color image, needs to be converted to numerical values before the actual analysis can be done. The analysis and information extraction of the raw data often remains the most challenging part for ChIP-on-chip experiments. Problems arise throughout this portion of the workflow, ranging from the initial chip read-out, to suitable methods to subtract background noise, and finally to appropriate
After a sufficiently large time frame to allow hybridization, the array is illuminated with fluorescent light. Those probes on the array that are hybridized to one of the labeled fragments emit a light signal that is captured by a camera. This image contains all raw data for the remaining part of the workflow.
This raw data, encoded as false-color image, needs to be converted to numerical values before the actual analysis can be done. The analysis and information extraction of the raw data often remains the most challenging part for ChIP-on-chip experiments. Problems arise throughout this portion of the workflow, ranging from the initial chip read-out, to suitable methods to subtract background noise, and finally to appropriate algorithm
In mathematics and computer science, an algorithm () is a finite sequence of Rigour#Mathematics, mathematically rigorous instructions, typically used to solve a class of specific Computational problem, problems or to perform a computation. Algo ...
s that normalize the data and make it available for subsequent statistical analysis
Statistical inference is the process of using data analysis to infer properties of an underlying probability distribution.Upton, G., Cook, I. (2008) ''Oxford Dictionary of Statistics'', OUP. . Inferential statistical analysis infers properties of ...
, which then hopefully lead to a better understanding of the biological question that the experiment seeks to address. Furthermore, due to the different array platforms and lack of standardization between them, data storage and exchange is a huge problem. Generally speaking, the data analysis can be divided into three major steps:
During the first step, the captured fluorescence signals from the array are normalized, using control signals derived from the same or a second chip. Such control signals tell which probes on the array were hybridized correctly and which bound nonspecifically.
In the second step, numerical and statistical tests are applied to control data and IP fraction data to identify POI-enriched regions along the genome. The following three methods are used widely: median percentile rank, single-array error, and sliding-window. These methods generally differ in how low-intensity signals are handled, how much background noise is accepted, and which trait for the data is emphasized during the computation. In the recent past, the sliding-window approach seems to be favored and is often described as most powerful.
In the third step, these regions are analyzed further. If, for example, the POI was a transcription factor, such regions would represent its binding sites. Subsequent analysis then may want to infer nucleotide motifs and other patterns to allow functional annotation of the genome.
Strengths and weaknesses
Using tiled arrays, ChIP-on-chip allows for high resolution of genome-wide maps. These maps can determine the binding sites of many DNA-binding proteins like transcription factors and also chromatin modifications. Although ChIP-on-chip can be a powerful technique in the area of genomics, it is very expensive. Most published studies using ChIP-on-chip repeat their experiments at least three times to ensure biologically meaningful maps. The cost of the DNA microarrays is often a limiting factor to whether a laboratory should proceed with a ChIP-on-chip experiment. Another limitation is the size of DNA fragments that can be achieved. Most ChIP-on-chip protocols utilize sonication as a method of breaking up DNA into small pieces. However, sonication is limited to a minimal fragment size of 200 bp. For higher resolution maps, this limitation should be overcome to achieve smaller fragments, preferably to singlenucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone, histone proteins and resembles thread wrapped around a bobbin, spool. The nucleosome ...
resolution. As mentioned previously, the statistical analysis of the huge amount of data generated from arrays is a challenge and normalization procedures should aim to minimize artifacts and determine what is really biologically significant. So far, application to mammalian genomes has been a major limitation, for example, due to the significant percentage of the genome that is occupied by repeats. However, as ChIP-on-chip technology advances, high resolution whole mammalian genome maps should become achievable.
Antibodies
An antibody (Ab) or immunoglobulin (Ig) is a large, Y-shaped protein belonging to the immunoglobulin superfamily which is used by the immune system to identify and neutralize antigens such as bacteria and viruses, including those that caus ...
used for ChIP-on-chip can be an important limiting factor. ChIP-on-chip requires highly specific antibodies that must recognize its epitope
An epitope, also known as antigenic determinant, is the part of an antigen that is recognized by the immune system, specifically by antibodies, B cells, or T cells. The part of an antibody that binds to the epitope is called a paratope. Although e ...
in free solution and also under fixed conditions. If it is demonstrated to successfully immunoprecipitate cross-linked chromatin
Chromatin is a complex of DNA and protein found in eukaryote, eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important r ...
, it is termed " ChIP-grade". Companies that provide ChIP-grade antibodies include Abcam, Cell Signaling Technology, Santa Cruz, and Upstate. To overcome the problem of specificity, the protein of interest can be fused to a tag like FLAG
A flag is a piece of textile, fabric (most often rectangular) with distinctive colours and design. It is used as a symbol, a signalling device, or for decoration. The term ''flag'' is also used to refer to the graphic design employed, and fla ...
or HA that are recognized by antibodies. An alternative to ChIP-on-chip that does not require antibodies is DamID.
Also available are antibodies against a specific histone modification like H3 tri methyl K4. As mentioned before, the combination of these antibodies and ChIP-on-chip has become extremely powerful in determining whole genome analysis of histone modification patterns and will contribute tremendously to our understanding of the histone code
The histone code is a hypothesis that the transcription of genetic information encoded in DNA is in part regulated by chemical modifications (known as ''histone marks'') to histone proteins, primarily on their unstructured ends. Together with sim ...
and epigenetics.
A study demonstrating the non-specific nature of DNA binding proteins has been published in PLoS Biology. This indicates that alternate confirmation of functional relevancy is a necessary step in any ChIP-chip experiment.
History
A first ChIP-on-chip experiment was performed in 1999 to analyze the distribution of cohesin alongbudding
Budding or blastogenesis is a type of asexual reproduction in which a new organism develops from an outgrowth or bud due to cell division at one particular site. For example, the small bulb-like projection coming out from the yeast cell is kno ...
yeast
Yeasts are eukaryotic, single-celled microorganisms classified as members of the fungus kingdom (biology), kingdom. The first yeast originated hundreds of millions of years ago, and at least 1,500 species are currently recognized. They are est ...
chromosome III. Although the genome was not completely represented, the protocol in this study remains equivalent as those used in later studies. The ChIP-on-chip technique using all of the ORFs of the genome (that nevertheless remains incomplete, missing intergenic regions) was then applied successfully in three papers published in 2000 and 2001. The authors identified binding sites for individual transcription factors in the budding
Budding or blastogenesis is a type of asexual reproduction in which a new organism develops from an outgrowth or bud due to cell division at one particular site. For example, the small bulb-like projection coming out from the yeast cell is kno ...
yeast
Yeasts are eukaryotic, single-celled microorganisms classified as members of the fungus kingdom (biology), kingdom. The first yeast originated hundreds of millions of years ago, and at least 1,500 species are currently recognized. They are est ...
''Saccharomyces cerevisiae
''Saccharomyces cerevisiae'' () (brewer's yeast or baker's yeast) is a species of yeast (single-celled fungal microorganisms). The species has been instrumental in winemaking, baking, and brewing since ancient times. It is believed to have be ...
''. In 2002, Richard Young's group determined the genome-wide positions of 106 transcription factors using a c-Myc tagging system in yeast. The first demonstration of the mammalian ChIp-on-chip technique reported the isolation of nine chromatin fragments containing weak and strong E2F binding site was done by Peggy Farnham's lab in collaboration with Michael Zhang's lab and published in 2001. This study was followed several months later in a collaboration between the Young lab with the laboratory of Brian Dynlacht which used the ChIP-on-chip technique to show for the first time that E2F targets encode components of the DNA damage checkpoint and repair pathways, as well as factors involved in chromatin assembly/condensation, chromosome segregation, and the mitotic spindle checkpoint Other applications for ChIP-on-chip include DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all life, living organisms, acting as the most essential part of heredity, biolog ...
, recombination, and chromatin structure. Since then, ChIP-on-chip has become a powerful tool in determining genome-wide maps of histone modifications and many more transcription factors. ChIP-on-chip in mammalian systems has been difficult due to the large and repetitive genomes. Thus, many studies in mammalian cells have focused on select promoter regions that are predicted to bind transcription factors and have not analyzed the entire genome. However, whole mammalian genome arrays have recently become commercially available from companies like Nimblegen. In the future, as ChIP-on-chip arrays become more and more advanced, high resolution whole genome maps of DNA-binding proteins and chromatin components for mammals will be analyzed in more detail.
Alternatives
Introduced in 2007, ChIP sequencing (ChIP-seq) is a technology that uses chromatin immunoprecipitation to crosslink the proteins of interest to the DNA but then instead of using a micro-array, it uses the more accurate, higher throughput method of sequencing to localize interaction points. DamID is an alternative method that does not require antibodies. ChIP-exo uses exonuclease treatment to achieve up to single base pair resolution. CUT&RUN sequencing uses antibody recognition with targeted enzymatic cleavage to address some technical limitations of ChIP.References
Further reading
* *External links
* http://www.genome.gov/10005107 ENCODE projectChip-on-Chip (CoC)
Package Information from Amkor Technology
Analysis and software
CoCAS: a free Analysis software for Agilent ChIP-on-Chip experiments
rMAT: R implementation from MAT program to normalize and analyze tiling arrays and ChIP-chip data. {{DEFAULTSORT:Chip-On-Chip Genomics techniques Molecular biology Molecular biology techniques Protein methods Bioinformatics Microarrays