|
Mitophagy
Mitophagy is the selective degradation of mitochondria by autophagy. It often occurs to defective mitochondria following damage or stress. The process of mitophagy was first described in 1915 by Margaret Reed Lewis and Warren Harmon Lewis. Ashford and Porter used electron microscopy to observe mitochondrial fragments in liver lysosomes by 1962, and a 1977 report suggested that "mitochondria develop functional alterations which would activate autophagy." The term "mitophagy" was coined by J.J. Lemasters et al. in 2005, though earlier uses dating back to at least 1998 can be found. Mitophagy is key in keeping the cell healthy. It promotes turnover of mitochondria and prevents accumulation of dysfunctional mitochondria which can lead to cellular degeneration. It is mediated by Atg32 (in yeast) and NIX and its regulator BNIP3 in mammals. Mitophagy is regulated by PINK1 and parkin proteins. In addition to the selective removal of damaged mitochondria, mitophagy is also required to ad ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Autophagy
Autophagy (or autophagocytosis; from the Greek language, Greek , , meaning "self-devouring" and , , meaning "hollow") is the natural, conserved degradation of the cell that removes unnecessary or dysfunctional components through a lysosome-dependent regulated mechanism. It allows the orderly degradation and recycling of cellular components. Although initially characterized as a primordial degradation pathway induced to protect against starvation, it has become increasingly clear that autophagy also plays a major role in the homeostasis of non-starved cells. Defects in autophagy have been linked to various human diseases, including neurodegeneration and cancer, and interest in modulating autophagy as a potential treatment for these diseases has grown rapidly. Four forms of autophagy have been identified: macroautophagy, microautophagy, chaperone-mediated autophagy (CMA), and crinophagy. In macroautophagy (the most thoroughly researched form of autophagy), cytoplasmic components ( ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Parkin (ligase)
Parkin is a 465-amino acid residue E3 ubiquitin ligase, a protein that in humans and mice is encoded by the ''PARK2'' gene. Parkin plays a critical role in ubiquitination – the process whereby molecules are covalently labelled with ubiquitin (Ub) and directed towards degradation in proteasomes or lysosomes. Ubiquitination involves the sequential action of three enzymes. First, an E1 ubiquitin-activating enzyme binds to inactive Ub in eukaryotic cells via a thioester bond and mobilises it in an ATP-dependent process. Ub is then transferred to an E2 ubiquitin-conjugating enzyme before being conjugated to the target protein via an E3 ubiquitin ligase. There exists a multitude of E3 ligases, which differ in structure and substrate specificity to allow selective targeting of proteins to intracellular degradation. In particular, parkin recognises proteins on the outer membrane of mitochondria upon cellular insult and mediates the clearance of damaged mitochondria via autophagy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nix (gene)
Nix is a pro-apoptotic gene that is regulated by Histotoxic hypoxia. It expresses a signaling protein related to the BH3-only family. This protein induces autophagy, an intracellular function by which cytoplasmic components are delivered to the lysosome to be broken down and used elsewhere or excreted from the cell. This protein is important in development because it allows cells to have a consistent store of cellular components. It also holds an important role in the differentiation and maturation of erythrocytes and lymphocytes by the process of mitophagy with the help of its regulator BNIP3. Using a gene knockout technique in mice, scientists have been able to attribute this pruning of mitochondria and induction of cellular necrosis to the expression of the Nix gene. The Nix protein may be associated with certain kinds of cancer formation. In mouse models, loss of Nix resulted in a delayed onset of tumors for pancreatic cancer, and was additionally associated with reduced mi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
BNIP3
BCL2/adenovirus E1B 19 kDa protein-interacting protein 3 is a protein found in humans that is encoded by the ''BNIP3'' gene. BNIP3 is a member of the apoptotic Bcl-2 protein family. It can induce cell death while also assisting with cell survival. Like many of the Bcl-2 family proteins, BNIP3 modulates the permeability state of the outer mitochondrial membrane by forming homo- and hetero-oligomers inside the membrane. Upregulation results in a decrease in mitochondrial potential, an increase in reactive oxygen species, mitochondrial swelling and fission, and an increase in mitochondrial turnover via autophagy. Sequence similarity with Bcl-2 family members was not detected. Humans and other animals (''Drosophila, Caenorhabditis''), as well as lower eukaryotes (''Dictyostelium, Trypanosoma, Cryptosporidium, Paramecium'') encode several BNIP3 paralogues including the human NIP3L, which induces apoptosis by interacting with viral and cellular anti-apoptosis proteins. Structure T ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
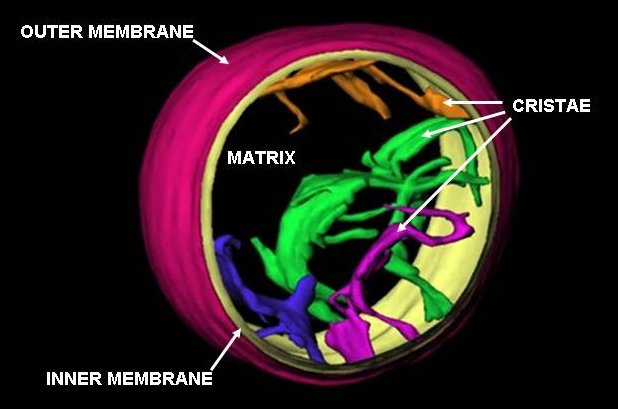
Mitochondria
A mitochondrion () is an organelle found in the cells of most eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used throughout the cell as a source of chemical energy. They were discovered by Albert von Kölliker in 1857 in the voluntary muscles of insects. The term ''mitochondrion'', meaning a thread-like granule, was coined by Carl Benda in 1898. The mitochondrion is popularly nicknamed the "powerhouse of the cell", a phrase popularized by Philip Siekevitz in a 1957 ''Scientific American'' article of the same name. Some cells in some multicellular organisms lack mitochondria (for example, mature mammalian red blood cells). The multicellular animal '' Henneguya salminicola'' is known to have retained mitochondrion-related organelles despite a complete loss of their mitochondrial genome. A large number of unicellular organisms, such as microspo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitylation
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ''ubiquitously''. It was discovered in 1975 by Gideon Goldstein and further characterized throughout the late 1970s and 1980s. Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A. The addition of ubiquitin to a substrate protein is called ubiquitylation (or ubiquitination or ubiquitinylation). Ubiquitylation affects proteins in many ways: it can mark them for degradation via the 26S proteasome, alter their cellular location, affect their activity, and promote or prevent protein interactions. Ubiquitylation involves three main steps: activation, conjugation, and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), respectively. The result of this sequential cascade is to bind ubiquitin to lysine residues on the protein substrate via an isopeptide bond, cystei ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MFN1
Mitofusin-1 is a protein that in humans is encoded by the ''MFN1'' gene. The protein encoded by this gene is a mediator of mitochondrial fusion. This protein and mitofusin 2 are homologs of the ''Drosophila ''Drosophila'' (), from Ancient Greek δρόσος (''drósos''), meaning "dew", and φίλος (''phílos''), meaning "loving", is a genus of fly, belonging to the family Drosophilidae, whose members are often called "small fruit flies" or p ...'' protein fuzzy onion (Fzo). They are mitochondrial membrane proteins that interact with each other to facilitate mitochondrial targeting. References Further reading * * * * * * * * * * * * * {{protein-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MFN2
Mitofusin-2 is a protein that in humans is encoded by the ''MFN2'' gene. Mitofusins are GTPases embedded in the outer membrane of the mitochondria. In mammals MFN1 and MFN2 are essential for mitochondrial fusion. In addition to the mitofusins, OPA1 regulates inner mitochondrial membrane fusion, and DRP1 is responsible for mitochondrial fission. Mitofusin-2 (MFN2) is a mitochondrial membrane protein that plays a central role in regulating mitochondrial fusion and cell metabolism. More specifically, MFN2 is a dynamin-like GTPase embedded in the outer mitochondrial membrane (OMM) which in turn affects mitochondrial dynamics, distribution, quality control, and function. In addition to the MFN2, OPA1 regulates inner mitochondrial membrane fusion, MFN1 is a mediator of mitochondrial fusion and DRP1 is responsible for mitochondrial fission. Structure The human mitofusin-2 protein contains 757 amino acid residues. The MFN2 comprises a large cytosolic GTPase domain at the N-term ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Senescence
Senescence () or biological aging is the gradual deterioration of Function (biology), functional characteristics in living organisms. Whole organism senescence involves an increase in mortality rate, death rates or a decrease in fecundity with increasing age, at least in the later part of an organism's biological life cycle, life cycle. However, the resulting effects of senescence can be delayed. The 1934 discovery that calorie restriction can Life extension, extend lifespans by 50% in rats, the existence of species having negligible senescence, and the existence of potentially immortal organisms such as members of the genus ''Hydra (genus), Hydra'' have motivated research into Life extension, delaying senescence and thus age-related diseases. Rare human mutations can cause accelerated aging diseases. Environmental Gerontogens, factors may affect aging – for example, overexposure to ultraviolet radiation accelerates skin aging. Different parts of the body may age at different ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MitoNEET
The CDGSH iron sulfur domain are a group of iron-sulfur (2Fe-2S) clusters and a unique 39 amino acid CDGSH domain ''C-X-C-X2-(S/T)-X3-P-X-C-D-G-(S/A/T)-H The CDGSH iron sulfur domain 1 protein (also referred to as mitoNEET) is an integral membrane protein located in the outer mitochondrial membrane and whose function may be to transport iron into the mitochondria. Iron in turn is essential for the function of several mitochondrial enzymes. The antidiabetic drug pioglitazone, in addition to binding to the nuclear receptor In the field of molecular biology, nuclear receptors are a class of proteins responsible for sensing steroids, thyroid hormones, vitamins, and certain other molecules. These intracellular receptors work with other proteins to regulate the ex ... PPAR, also has been shown to bind mitoNEET with approximately equal affinity. References External links * {{MeshName, ZCD1+protein,+human Protein domains ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ubiquitin Ligase
A ubiquitin ligase (also called an E3 ubiquitin ligase) is a protein that recruits an E2 ubiquitin-conjugating enzyme that has been loaded with ubiquitin, recognizes a protein substrate, and assists or directly catalyzes the transfer of ubiquitin from the E2 to the protein substrate. In simple and more general terms, the ligase enables movement of ubiquitin from a ubiquitin carrier to another protein (the substrate) by some mechanism. The ubiquitin, once it reaches its destination, ends up being attached by an isopeptide bond to a lysine residue, which is part of the target protein. E3 ligases interact with both the target protein and the E2 enzyme, and so impart substrate specificity to the E2. Commonly, E3s polyubiquitinate their substrate with Lys48-linked chains of ubiquitin, targeting the substrate for destruction by the proteasome. However, many other types of linkages are possible and alter a protein's activity, interactions, or localization. Ubiquitination by E3 ligases re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |