Variants of PCR on:
[Wikipedia]
[Google]
[Amazon]
Variants of PCR represent a diverse array of techniques that have evolved from the basic
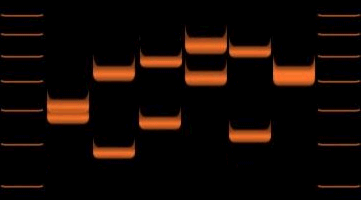 ''Variable Number of Tandem Repeats (VNTR) PCR'' targets repetitive areas of the genome that exhibit length variation. Analysis of the genotypes in the samples usually involves sizing of the amplification products by gel electrophoresis. Analysis of smaller VNTR segments known as short tandem repeats (or STRs) is the basis for DNA fingerprinting
''Variable Number of Tandem Repeats (VNTR) PCR'' targets repetitive areas of the genome that exhibit length variation. Analysis of the genotypes in the samples usually involves sizing of the amplification products by gel electrophoresis. Analysis of smaller VNTR segments known as short tandem repeats (or STRs) is the basis for DNA fingerprinting  '' Nested PCR'' is used to increase the specificity of DNA amplification. Two sets of primers are used in two successive reactions. In the first PCR, one pair of primers is used to generate DNA products, which may contain products amplified from non-target areas. The products from the first PCR are then used as template in a second PCR, using one ('hemi-nesting') or two different primers whose binding sites are located (nested) within the first set, thus increasing specificity. Nested PCR is often more successful in specifically amplifying long DNA products than conventional PCR, but it requires more detailed knowledge of the sequence of the target.
'' Quantitative PCR'' (''qPCR'') is used to measure the specific amount of target DNA (or RNA) in a sample. By measuring amplification only within the phase of true exponential increase, the amount of measured product more accurately reflects the initial amount of target. Special thermal cyclers are used that monitor the amount of product during the amplification.
''Quantitative Real-Time PCR'' (''QRT-PCR''), sometimes simply called ''Real-Time PCR'' (''RT-PCR''), refers to a collection of methods that use fluorescent dyes, such as Sybr Green, or fluorophore-containing DNA probes, such as TaqMan, to measure the amount of amplified product in real time as the amplification progresses.
''Hot-start PCR'' is a technique performed manually by heating the reaction components to the DNA melting temperature (e.g. 95 °C) before adding the polymerase. In this way, non-specific amplification at lower temperatures is prevented. Alternatively, specialized reagents inhibit the polymerase's activity at ambient temperature, either by the binding of an
'' Nested PCR'' is used to increase the specificity of DNA amplification. Two sets of primers are used in two successive reactions. In the first PCR, one pair of primers is used to generate DNA products, which may contain products amplified from non-target areas. The products from the first PCR are then used as template in a second PCR, using one ('hemi-nesting') or two different primers whose binding sites are located (nested) within the first set, thus increasing specificity. Nested PCR is often more successful in specifically amplifying long DNA products than conventional PCR, but it requires more detailed knowledge of the sequence of the target.
'' Quantitative PCR'' (''qPCR'') is used to measure the specific amount of target DNA (or RNA) in a sample. By measuring amplification only within the phase of true exponential increase, the amount of measured product more accurately reflects the initial amount of target. Special thermal cyclers are used that monitor the amount of product during the amplification.
''Quantitative Real-Time PCR'' (''QRT-PCR''), sometimes simply called ''Real-Time PCR'' (''RT-PCR''), refers to a collection of methods that use fluorescent dyes, such as Sybr Green, or fluorophore-containing DNA probes, such as TaqMan, to measure the amount of amplified product in real time as the amplification progresses.
''Hot-start PCR'' is a technique performed manually by heating the reaction components to the DNA melting temperature (e.g. 95 °C) before adding the polymerase. In this way, non-specific amplification at lower temperatures is prevented. Alternatively, specialized reagents inhibit the polymerase's activity at ambient temperature, either by the binding of an
 Unlike normal PCR, '' Inverse PCR'' allows amplification and sequencing of DNA that surrounds a known sequence. It involves initially subjecting the target DNA to a series of
Unlike normal PCR, '' Inverse PCR'' allows amplification and sequencing of DNA that surrounds a known sequence. It involves initially subjecting the target DNA to a series of
PCR Applications Manual (from Roche Diagnostics).
The Reference in qPCR -- an Academic & Industrial Information Platform
www.eConferences.de streaming portal -- Amplify your knowledge in qPCR, dPCR and NGS!
{{DEFAULTSORT:Variants Of Pcr Polymerase chain reaction
polymerase chain reaction
The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA (or a part of it) sufficiently to enable detailed st ...
(PCR) method, each tailored to specific applications in molecular biology
Molecular biology is a branch of biology that seeks to understand the molecule, molecular basis of biological activity in and between Cell (biology), cells, including biomolecule, biomolecular synthesis, modification, mechanisms, and interactio ...
, such as genetic analysis, DNA sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, thymine, cytosine, and guanine. The ...
, and disease diagnosis, by modifying factors like primer design, temperature conditions, and enzyme usage.
Basic modifications
Often only a small modification needs to be made to the standard PCR protocol to achieve a desired goal: '' Multiplex-PCR'' uses several pairs of primers annealing to different target sequences. This permits the simultaneous analysis of multiple targets in a single sample. For example, in testing for genetic mutations, six or more amplifications might be combined. In the standard protocol for DNA fingerprinting, the targets assayed are often amplified in groups of 3 or 4. ''Multiplex Ligation-dependent Probe Amplification
Multiplex ligation-dependent probe amplification (MLPA) is a variation of the multiplex polymerase chain reaction that permits amplification of multiple targets with only a single primer (molecular biology), primer pair. It detects copy number cha ...
'' (''MLPA'') permits multiple targets to be amplified using only a single pair of primers, avoiding the resolution limitations of multiplex PCR. Multiplex PCR has also been used for analysis of microsatellites and SNPs.
 ''Variable Number of Tandem Repeats (VNTR) PCR'' targets repetitive areas of the genome that exhibit length variation. Analysis of the genotypes in the samples usually involves sizing of the amplification products by gel electrophoresis. Analysis of smaller VNTR segments known as short tandem repeats (or STRs) is the basis for DNA fingerprinting
''Variable Number of Tandem Repeats (VNTR) PCR'' targets repetitive areas of the genome that exhibit length variation. Analysis of the genotypes in the samples usually involves sizing of the amplification products by gel electrophoresis. Analysis of smaller VNTR segments known as short tandem repeats (or STRs) is the basis for DNA fingerprinting databases
In computing, a database is an organized collection of data or a type of data store based on the use of a database management system (DBMS), the software that interacts with end users, applications, and the database itself to capture and ana ...
such as CODIS.
''Asymmetric PCR'' preferentially amplifies one strand of a double-stranded DNA target. It is used in some sequencing
In genetics and biochemistry, sequencing means to determine the primary structure (sometimes incorrectly called the primary sequence) of an unbranched biopolymer. Sequencing results in a symbolic linear depiction known as a sequence which succ ...
methods and hybridization probing to generate one DNA strand as product. Thermocycling is carried out exactly as in conventional PCR, but with a limiting amount or leaving out one of the primers. When the limiting primer becomes depleted, replication increases arithmetically rather than exponentially through extension of the excess primer. A modification of this process, named ''L''inear-''A''fter-''T''he-''E''xponential-PCR (or ''LATE-PCR''), uses a limiting primer with a higher melting temperature (Tm) than the excess primer in order to maintain reaction efficiency as the limiting primer concentration decreases mid-reaction. See also overlap-extension PCR.
Some modifications are needed to perform ''long PCR''. The original Klenow-based PCR process did not generate products that were larger than about 400 bp. Taq polymerase can however amplify targets of up to several thousand bp long. Since then, modified protocols with Taq enzyme have allowed targets of over 50 kb to be amplified.
 '' Nested PCR'' is used to increase the specificity of DNA amplification. Two sets of primers are used in two successive reactions. In the first PCR, one pair of primers is used to generate DNA products, which may contain products amplified from non-target areas. The products from the first PCR are then used as template in a second PCR, using one ('hemi-nesting') or two different primers whose binding sites are located (nested) within the first set, thus increasing specificity. Nested PCR is often more successful in specifically amplifying long DNA products than conventional PCR, but it requires more detailed knowledge of the sequence of the target.
'' Quantitative PCR'' (''qPCR'') is used to measure the specific amount of target DNA (or RNA) in a sample. By measuring amplification only within the phase of true exponential increase, the amount of measured product more accurately reflects the initial amount of target. Special thermal cyclers are used that monitor the amount of product during the amplification.
''Quantitative Real-Time PCR'' (''QRT-PCR''), sometimes simply called ''Real-Time PCR'' (''RT-PCR''), refers to a collection of methods that use fluorescent dyes, such as Sybr Green, or fluorophore-containing DNA probes, such as TaqMan, to measure the amount of amplified product in real time as the amplification progresses.
''Hot-start PCR'' is a technique performed manually by heating the reaction components to the DNA melting temperature (e.g. 95 °C) before adding the polymerase. In this way, non-specific amplification at lower temperatures is prevented. Alternatively, specialized reagents inhibit the polymerase's activity at ambient temperature, either by the binding of an
'' Nested PCR'' is used to increase the specificity of DNA amplification. Two sets of primers are used in two successive reactions. In the first PCR, one pair of primers is used to generate DNA products, which may contain products amplified from non-target areas. The products from the first PCR are then used as template in a second PCR, using one ('hemi-nesting') or two different primers whose binding sites are located (nested) within the first set, thus increasing specificity. Nested PCR is often more successful in specifically amplifying long DNA products than conventional PCR, but it requires more detailed knowledge of the sequence of the target.
'' Quantitative PCR'' (''qPCR'') is used to measure the specific amount of target DNA (or RNA) in a sample. By measuring amplification only within the phase of true exponential increase, the amount of measured product more accurately reflects the initial amount of target. Special thermal cyclers are used that monitor the amount of product during the amplification.
''Quantitative Real-Time PCR'' (''QRT-PCR''), sometimes simply called ''Real-Time PCR'' (''RT-PCR''), refers to a collection of methods that use fluorescent dyes, such as Sybr Green, or fluorophore-containing DNA probes, such as TaqMan, to measure the amount of amplified product in real time as the amplification progresses.
''Hot-start PCR'' is a technique performed manually by heating the reaction components to the DNA melting temperature (e.g. 95 °C) before adding the polymerase. In this way, non-specific amplification at lower temperatures is prevented. Alternatively, specialized reagents inhibit the polymerase's activity at ambient temperature, either by the binding of an antibody
An antibody (Ab) or immunoglobulin (Ig) is a large, Y-shaped protein belonging to the immunoglobulin superfamily which is used by the immune system to identify and neutralize antigens such as pathogenic bacteria, bacteria and viruses, includin ...
, or by the presence of covalently bound inhibitors that only dissociate after a high-temperature activation step. 'Hot-start/cold-finish PCR' is achieved with new hybrid polymerases that are inactive at ambient temperature and are only activated at elevated temperatures.
In '' touchdown PCR'', the annealing temperature is gradually decreased in later cycles. The annealing temperature in the early cycles is usually 3–5 °C above the standard Tm of the primers used, while in the later cycles it is a similar amount below the Tm. The initial higher annealing temperature leads to greater specificity for primer binding, while the lower temperatures permit more efficient amplification at the end of the reaction.
'' Assembly PCR'' (also known as ''Polymerase Cycling Assembly'' or ''PCA'') is the synthesis of long DNA structures by performing PCR on a pool of long oligonucleotides with short overlapping segments, to assemble two or more pieces of DNA into one piece. It involves an initial PCR with primers that have an overlap and a second PCR using the products as the template that generates the final full-length product. This technique may substitute for ligation-based assembly.
In ''colony PCR'', bacterial colonies are screened directly by PCR, for example, the screen for correct DNA vector
Vector most often refers to:
* Euclidean vector, a quantity with a magnitude and a direction
* Disease vector, an agent that carries and transmits an infectious pathogen into another living organism
Vector may also refer to:
Mathematics a ...
constructs. Colonies are sampled with a sterile pipette tip and a small quantity of cells transferred into a PCR mix. To release the DNA from the cells, the PCR is either started with an extended time at 95 °C (when standard polymerase is used), or with a shortened denaturation step at 100 °C and special chimeric DNA polymerase.
The '' digital polymerase chain reaction'' simultaneously amplifies thousands of samples, each in a separate droplet within an emulsion or partition within an micro-well.
''Suicide PCR'' is typically used in paleogenetics or other studies where avoiding false positives and ensuring the specificity of the amplified fragment is the highest priority. It was originally described in a study to verify the presence of the microbe '' Yersinia pestis'' in dental samples obtained from 14th-century graves of people supposedly killed by plague during the medieval Black Death
The Black Death was a bubonic plague pandemic that occurred in Europe from 1346 to 1353. It was one of the list of epidemics, most fatal pandemics in human history; as many as people perished, perhaps 50% of Europe's 14th century population. ...
epidemic. The method prescribes the use of any primer combination only once in a PCR (hence the term "suicide"), which should never have been used in any positive-control PCR reaction, and the primers should always target a genomic region never amplified before in the lab using this or any other set of primers. This ensures that no contaminating DNA from previous PCR reactions is present in the lab, which could otherwise generate false positives.
COLD-PCR (''co''-amplification at ''l''ower ''d''enaturation temperature-PCR) is a modified protocol that enriches variant alleles from a mixture of wild-type and mutation-containing DNA samples.
Pretreatments and extensions
The basic PCR process can sometimes precede or follow another technique. '' RT-PCR'' (or ''R''everse ''T''ranscription ''PCR'') is used to reverse-transcribe and amplifyRNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA and deoxyrib ...
to cDNA. PCR is preceded by a reaction using reverse transcriptase
A reverse transcriptase (RT) is an enzyme used to convert RNA genome to DNA, a process termed reverse transcription. Reverse transcriptases are used by viruses such as HIV and hepatitis B to replicate their genomes, by retrotransposon mobi ...
, an enzyme that converts RNA into cDNA. The two reactions may be combined in a tube, with the initial heating step of PCR being used to inactivate the transcriptase. The Tth polymerase (described below) has RT activity, and can carry out the entire reaction. RT-PCR is widely used in expression profiling, which detects the expression of a gene. It can also be used to obtain sequence of an RNA transcript, which may aid the determination of the transcription start and termination sites (by RACE-PCR) and facilitate mapping of the location of exons
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequence i ...
and introns
An intron is any Nucleic acid sequence, nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e., a region inside a gene."The notion of ...
in a gene sequence. Multiplex qRT-PCR has been developed to enhance SARS-CoV-2 detection efficiency by allowing multiple viral targets to be tested simultaneously.
''Two-tailed PCR'' uses a single primer that binds to a microRNA target with both 3' and 5' ends, known as hemiprobes. Both ends must be complementary for binding to occur. The 3'-end is then extended by reverse transcriptase forming a long cDNA. The cDNA is then amplified using two target specific PCR primers. The combination of two hemiprobes, both targeting the short microRNA target, makes the Two-tailed assay exceedingly sensitive and specific.
''Ligation-mediated PCR'' uses small DNA oligonucleotide 'linkers' (or adaptors) that are first ligated to fragments of the target DNA. PCR primers that anneal to the linker sequences are then used to amplify the target fragments. This method is deployed for DNA sequencing, genome walking, and DNA footprinting. A related technique is '' amplified fragment length polymorphism'', which generates diagnostic fragments of a genome.
''Methylation-specific PCR'' (''MSP'') is used to identify patterns of DNA methylation at cytosine-guanine (CpG) islands in genomic DNA. Target DNA is first treated with sodium bisulfite, which converts unmethylated cytosine
Cytosine () (symbol C or Cyt) is one of the four nucleotide bases found in DNA and RNA, along with adenine, guanine, and thymine ( uracil in RNA). It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attac ...
bases to uracil
Uracil () (nucleoside#List of nucleosides and corresponding nucleobases, symbol U or Ura) is one of the four nucleotide bases in the nucleic acid RNA. The others are adenine (A), cytosine (C), and guanine (G). In RNA, uracil binds to adenine via ...
, which is complementary to adenosine
Adenosine (symbol A) is an organic compound that occurs widely in nature in the form of diverse derivatives. The molecule consists of an adenine attached to a ribose via a β-N9- glycosidic bond. Adenosine is one of the four nucleoside build ...
in PCR primers. Two amplifications are then carried out on the bisulfite-treated DNA: one primer set anneals to DNA with cytosines (corresponding to methylated cytosine), and the other set anneals to DNA with uracil (corresponding to unmethylated cytosine). MSP used in quantitative PCR provides quantitative information about the methylation state of a given CpG island.
Other modifications
Adjustments of the components in PCR is commonly used for optimal performance. The divalent ''magnesium
Magnesium is a chemical element; it has Symbol (chemistry), symbol Mg and atomic number 12. It is a shiny gray metal having a low density, low melting point and high chemical reactivity. Like the other alkaline earth metals (group 2 ...
'' ion (Mg++) is required for PCR polymerase activity. Lower concentrations Mg++ will increase replication fidelity, while higher concentrations will introduce more mutations.
''Denaturants'' (such as dimethylsulfoxide) can increase amplification specificity by destabilizing non-specific primer binding. Other chemicals, such as glycerol
Glycerol () is a simple triol compound. It is a colorless, odorless, sweet-tasting, viscous liquid. The glycerol backbone is found in lipids known as glycerides. It is also widely used as a sweetener in the food industry and as a humectant in pha ...
, are ''stabilizers'' for the activity of the polymerase during amplification. ''Detergents ''(such as Triton X-100) can prevent polymerase stick to itself or to the walls of the reaction tube.
DNA polymerases occasionally incorporate mismatch bases into the extending strand. ''High-fidelity PCR'' employs enzymes with 3'-5' exonuclease activity that decreases this rate of mis-incorporation. Examples of enzymes with proofreading activity include Pfu; adjustments of the Mg++ and dNTP concentrations may help maximize the number of products that exactly match the original target DNA.
Primer modifications
Adjustments to the synthetic oligonucleotides used as primers in PCR are a rich source of modification: Normally PCR primers are chosen from an invariant part of the genome, and might be used to amplify a polymorphic area between them. In ''allele-specific PCR'' the opposite is done. At least one of the primers is chosen from a polymorphic area, with the mutations located at (or near) its 3'-end. Under stringent conditions, a mismatched primer will not initiate replication, whereas a matched primer will. The appearance of an amplification product therefore indicates the genotype. ''(For more information, seeSNP genotyping
SNP genotyping is the measurement of genetic variations of single nucleotide polymorphisms (SNPs) between members of a species. It is a form of genotyping, which is the measurement of more general genetic variation. SNPs are one of the most commo ...
.)''
''InterSequence-Specific PCR'' (or ''ISSR-PCR'') is method for DNA fingerprinting that uses primers selected from segments repeated throughout a genome to produce a unique fingerprint of amplified product lengths. The use of primers from a commonly repeated segment is called ''Alu-PCR'', and can help amplify sequences adjacent (or between) these repeats.
Primers can also be designed to be 'degenerate' – able to initiate replication from a large number of target locations. '' Whole genome amplification'' (or ''WGA'') is a group of procedures that allow amplification to occur at many locations in an unknown genome, and which may only be available in small quantities. Other techniques use ''degenerate primers'' that are synthesized using multiple nucleotides at particular positions (the polymerase 'chooses' the correctly matched primers). Also, the primers can be synthesized with the nucleoside analog inosine, which hybridizes to three of the four normal bases. A similar technique can force PCR to perform Site-directed mutagenesis. (also see Overlap extension polymerase chain reaction)
Normally the primers used in PCR are designed to be fully complementary to the target. However, the polymerase is tolerant to mis-matches away from the 3' end. ''Tailed-primers'' include non-complementary sequences at their 5' ends. A common procedure is the use of ''linker-primers'', which ultimately place restriction sites at the ends of the PCR products, facilitating their later insertion into cloning vectors.
An extension of the 'colony-PCR' method (above), is the use of ''vector primers''. Target DNA fragments (or cDNA) are first inserted into a cloning vector
Vector most often refers to:
* Euclidean vector, a quantity with a magnitude and a direction
* Disease vector, an agent that carries and transmits an infectious pathogen into another living organism
Vector may also refer to:
Mathematics a ...
, and a single set of primers are designed for the areas of the vector flanking the insertion site. Amplification occurs for whatever DNA has been inserted.
PCR can easily be modified to produce a ''labeled product'' for subsequent use as a hybridization probe. One or both primers might be used in PCR with a radioactive or fluorescent label already attached, or labels might be added after amplification. These labeling methods can be combined with 'asymmetric-PCR' (above) to produce effective hybridization probes.
'' RNase H-dependent PCR'' (rhPCR) can reduce primer-dimer formation, and increase the number of assays in multiplex PCR. The method utilizes primers with a cleavable block on the 3’ end that is removed by the action of a thermostable RNase HII enzyme.
DNA Polymerases
There are several DNA polymerases that are used in PCR. The '' Klenow fragment'', derived from the original DNA Polymerase I from ''E. coli'', was the first enzyme used in PCR. Because of its lack of stability at high temperature, it needs be replenished during each cycle, and therefore is not commonly used in PCR. The bacteriophage ''T4 DNA polymerase'' (family A) was also initially used in PCR. It has a higher fidelity of replication than the Klenow fragment, but is also destroyed by heat. T7 DNA polymerase (family B) has similar properties and purposes. It has been applied to site-directed mutagenesis andSanger sequencing
Sanger sequencing is a method of DNA sequencing that involves electrophoresis and is based on the random incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication. After first being developed by Fred ...
.
'' Taq'' polymerase, the DNA Polymerase I from ''Thermus aquaticus
''Thermus aquaticus'' is a species of bacteria that can tolerate high temperatures, one of several thermophile, thermophilic bacteria that belong to the ''Deinococcota'' phylum. It is the source of the heat-resistant enzyme Taq polymerase, ''Taq' ...
'', was the first thermostable polymerase used in PCR, and is still the one most commonly used. The enzyme can be isolated from its native source, or from its cloned gene expressed in ''E. coli''. A 61kDa truncated from lacking 5'-3' exonuclease activity is known as the ''Stoffel fragment'', and is expressed in ''E. coli''. The lack of exonuclease activity may allow it to amplify longer targets than the native enzyme. It has been commercialized as ''AmpliTaq'' and ''Klentaq''. A variant designed for hot-start PCR called the "Faststart polymerase" has also been produced. It requires strong heat activation, thereby avoiding non-specific amplification due to polymerase activity at low temperature. Many other variants have been created.
Other ''Thermus'' polymerases, such as ''Tth'' polymerase I () from '' Thermus thermophilus'', has seen some use. ''Tth'' has reverse transcriptase activity in the presence of Mn2+ ions, allowing PCR amplification from RNA targets.
The archean
The Archean ( , also spelled Archaean or Archæan), in older sources sometimes called the Archaeozoic, is the second of the four geologic eons of Earth's history of Earth, history, preceded by the Hadean Eon and followed by the Proterozoic and t ...
genus '' Pyrococcus'' has proven a rich source of thermostable polymerases with proofreading activity. '' Pfu DNA polymerase'', isolated from the '' P. furiosus'' shows a 5-fold decrease in the error rate of replication compared to Taq. Since errors increase as PCR progresses, Pfu is the preferred polymerase when products are to be individually cloned for sequencing or expression. Other lesser used polymerases from this genus include ''Pwo'' () from ''Pyrococcus woesei'', ''Pfx'' from an unnamed species, "Deep Vent" polymerase () from strain GB-D.
Vent or '' Tli polymerase'' is an extremely thermostable DNA polymerase isolated from '' Thermococcus litoralis''. The polymerase from '' Thermococcus fumicolans'' (''Tfu'') has also been commercialized.
Mechanism modifications
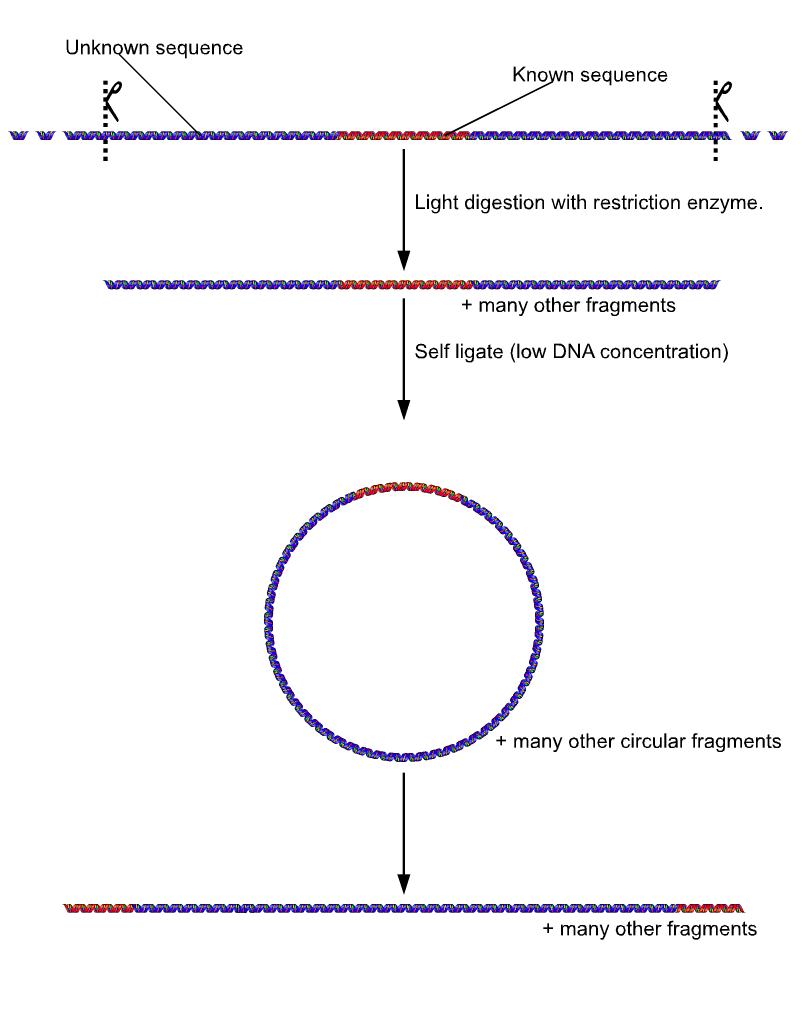
Sometimes even the basic mechanism of PCR can be modified. Unlike normal PCR, '' Inverse PCR'' allows amplification and sequencing of DNA that surrounds a known sequence. It involves initially subjecting the target DNA to a series of
Unlike normal PCR, '' Inverse PCR'' allows amplification and sequencing of DNA that surrounds a known sequence. It involves initially subjecting the target DNA to a series of restriction enzyme
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class o ...
digestions, and then circularizing the resulting fragments by self ligation. Primers are designed to be extended outward from the known segment, resulting in amplification of the rest of the circle. This is especially useful in identifying sequences to either side of various genomic inserts.
Similarly, ''thermal asymmetric interlaced PCR'' (or ''TAIL-PCR'') is used to isolate unknown sequences flanking a known area of the genome. Within the known sequence, TAIL-PCR uses a nested pair of primers with differing annealing temperatures. A 'degenerate' primer is used to amplify in the other direction from the unknown sequence.
Isothermal amplification methods
Some DNA amplification protocols have been developed that may be used alternatively to PCR. They are isothermal, meaning that they are run at a constant temperature. '' Helicase-dependent amplification'' (HDA) is similar to traditional PCR, but uses a constant temperature rather than cycling through denaturation and annealing/extension steps. DNA Helicase, an enzyme that unwinds DNA, is used in place of thermal denaturation. '' Loop-mediated isothermal amplification'' is a similar idea, but done with a strand-displacing polymerase. ''Nicking enzyme amplification reaction Nicking Enzyme Amplification Reaction (NEAR) is a method for '' in vitro'' DNA amplification like the polymerase chain reaction (PCR). NEAR is isothermal, replicating DNA at a constant temperature using a polymerase (and nicking enzyme) to expo ...
'' (NEAR) and its cousin strand displacement amplification (SDA) are isothermal, replicating DNA at a constant temperature using a polymerase and nicking enzyme.
'' Recombinase Polymerase Amplification'' (RPA) uses a recombinase to specifically pair primers with double-stranded DNA on the basis of homology, thus directing DNA synthesis from defined DNA sequences present in the sample. Presence of the target sequence initiates DNA amplification, and no thermal or chemical melting of DNA is required. The reaction progresses rapidly and results in specific DNA amplification from just a few target copies to detectable levels typically within 5–10 minutes. The entire reaction system is stable as a dried formulation and does not need refrigeration. RPA can be used to replace PCR in a variety of laboratory applications and users can design their own assays.
Other types of isothermal amplification include whole genome amplification (WGA), Nucleic acid sequence-based amplification (NASBA), and transcription-mediated amplification
Transcription-mediated amplification (TMA) is an isothermal (performed at constant temperature), single-tube nucleic acid Gene_duplication#As_amplification, amplification system utilizing two enzymes, RNA polymerase and reverse transcriptase.
"Am ...
(TMA).
See also
* Vectorette PCRReferences
External links
PCR Applications Manual (from Roche Diagnostics).
The Reference in qPCR -- an Academic & Industrial Information Platform
www.eConferences.de streaming portal -- Amplify your knowledge in qPCR, dPCR and NGS!
{{DEFAULTSORT:Variants Of Pcr Polymerase chain reaction