|
DNA Fingerprinting
DNA profiling (also called DNA fingerprinting) is the process of determining an individual's DNA characteristics. DNA analysis intended to identify a species, rather than an individual, is called DNA barcoding. DNA profiling is a forensic technique in criminal investigations, comparing criminal suspects' profiles to DNA evidence so as to assess the likelihood of their involvement in the crime. It is also used in paternity testing, to establish immigration eligibility, and in genealogical and medical research. DNA profiling has also been used in the study of animal and plant populations in the fields of zoology, botany, and agriculture. Background Starting in the 1980s, scientific advances allowed the use of DNA as a material for the identification of an individual. The first patent covering the direct use of DNA variation for forensicsUS5593832A was filed by Jeffrey Glassberg in 1983, based upon work he had done while at Rockefeller University in the United States in 1981. B ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
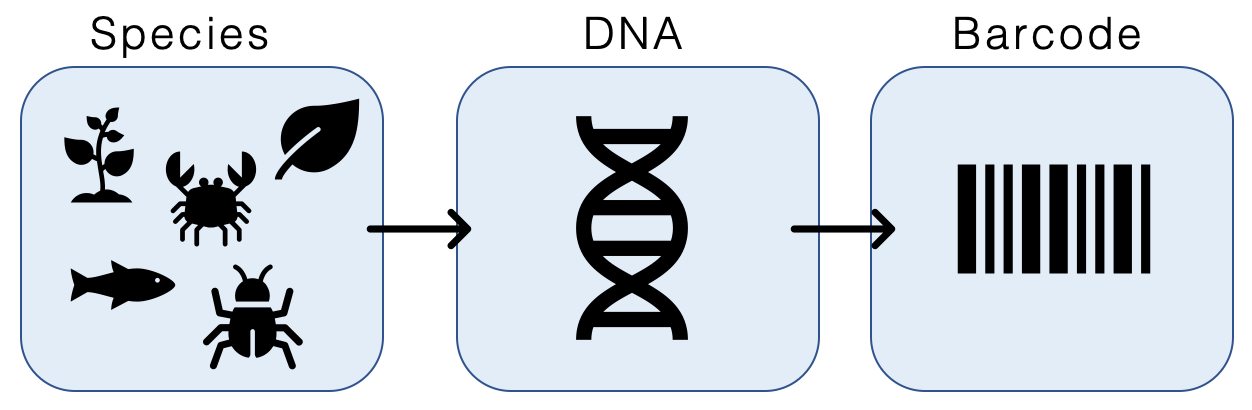
DNA Barcoding
DNA barcoding is a method of species identification using a short section of DNA from a specific gene or genes. The premise of DNA barcoding is that by comparison with a reference library of such DNA sections (also called "sequences"), an individual sequence can be used to uniquely identify an organism to species, just as a supermarket scanner uses the familiar black stripes of the UPC barcode to identify an item in its stock against its reference database. These "barcodes" are sometimes used in an effort to identify unknown species or parts of an organism, simply to catalog as many taxa as possible, or to compare with traditional taxonomy in an effort to determine species boundaries. Different gene regions are used to identify the different organismal groups using barcoding. The most commonly used barcode region for animals and some protists is a portion of the cytochrome ''c'' oxidase I (COI or COX1) gene, found in mitochondrial DNA. Other genes suitable for DNA barcoding ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Monozygotic Twins
Twins are two offspring produced by the same pregnancy.MedicineNet > Definition of TwinLast Editorial Review: 19 June 2000 Twins can be either ''monozygotic'' ('identical'), meaning that they develop from one zygote, which splits and forms two embryos, or ''dizygotic'' ('non-identical' or 'fraternal'), meaning that each twin develops from a separate egg and each egg is fertilized by its own sperm cell. Since identical twins develop from one zygote, they will share the same sex, while fraternal twins may or may not. In rare cases twins can have the same mother and different fathers (heteropaternal superfecundation). In contrast, a fetus that develops alone in the womb (the much more common case, in humans) is called a ''singleton'', and the general term for one offspring of a multiple birth is a ''multiple''. Unrelated look-alikes whose resemblance parallels that of twins are referred to as doppelgängers. Statistics The human twin birth rate in the United States rose 76% from ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
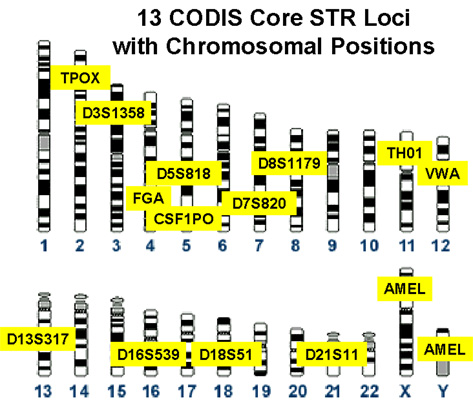
CODIS
The Combined DNA Index System (CODIS) is the United States national DNA database created and maintained by the Federal Bureau of Investigation. CODIS consists of three levels of information; Local DNA Index Systems (LDIS) where DNA profiles originate, State DNA Index Systems (SDIS) which allows for laboratories within states to share information, and the National DNA Index System (NDIS) which allows states to compare DNA information with one another. The CODIS software contains multiple different databases depending on the type of information being searched against. Examples of these databases include, missing persons, convicted offenders, and forensic samples collected from crime scenes. Each state, and the federal system, has different laws for collection, upload, and analysis of information contained within their database. However, for privacy reasons, the CODIS database does not contain any personal identifying information, such as the name associated with the DNA profile. Th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is a method widely used to rapidly make millions to billions of copies (complete or partial) of a specific DNA sample, allowing scientists to take a very small sample of DNA and amplify it (or a part of it) to a large enough amount to study in detail. PCR was invented in 1983 by the American biochemist Kary Mullis at Cetus Corporation; Mullis and biochemist Michael Smith (chemist), Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993. PCR is fundamental to many of the procedures used in genetic testing and research, including analysis of Ancient DNA, ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory research for a broad variety of applications ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Short Tandem Repeat (STR) Analysis
A microsatellite is a tract of repetitive DNA in which certain DNA motifs (ranging in length from one to six or more base pairs) are repeated, typically 5–50 times. Microsatellites occur at thousands of locations within an organism's genome. They have a higher mutation rate than other areas of DNA leading to high genetic diversity. Microsatellites are often referred to as short tandem repeats (STRs) by forensic geneticists and in genetic genealogy, or as simple sequence repeats (SSRs) by plant geneticists. Microsatellites and their longer cousins, the minisatellites, together are classified as VNTR (variable number of tandem repeats) DNA. The name "satellite" DNA refers to the early observation that centrifugation of genomic DNA in a test tube separates a prominent layer of bulk DNA from accompanying "satellite" layers of repetitive DNA. They are widely used for DNA profiling in cancer diagnosis, in kinship analysis (especially paternity testing) and in forensic identific ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Differential Extraction
Differential extraction (also known as differential lysis) refers to the process by which the DNA from two different types of cells can be extracted without mixing their contents. The most common application of this method is the extraction of DNA from vaginal epithelial cells and sperm cells from sexual assault cases in order to determine the DNA profiles of the victim and the perpetrator. Its success is based on the fact that sperm cells pack their DNA using protamines (rather than histones) which are held together by disulfide bonds. The protamines sequester DNA from spermatozoa, making it more resilient to DNA extraction than DNA from epithelial cells. After determining that sperm cells are present (typically through staining and light microscopy) in a vaginal/rectal sample, the subject's epithelial cells are lysed by a standard DNA extraction method, like a phenol/chloroform extraction and their DNA extracted through normal means. The epithelial DNA in solution is removed and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Solid Phase Extraction
Solid-phase extraction (SPE) is an extractive technique by which compounds that are dissolved or suspended in a liquid mixture are separated from other compounds in the mixture according to their physical and chemical properties. Analytical laboratories use solid phase extraction to concentrate and purify samples for analysis. Solid phase extraction can be used to isolate analytes of interest from a wide variety of matrices, including urine, blood, water, beverages, soil, and animal tissue. SPE uses the affinity of solutes dissolved or suspended in a liquid (known as the mobile phase) for a solid through which the sample is passed (known as the stationary phase) to separate a mixture into desired and undesired components. The result is that either the desired analytes of interest or undesired impurities in the sample are retained on the stationary phase. The portion that passes through the stationary phase is collected or discarded, depending on whether it contains the desired ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chelex 100
Chelex 100 is a chelating material from Bio-Rad used to purify other compounds via ion exchange. It is noteworthy for its ability to bind transition metal ions. It is a styrene-divinylbenzene co-polymer containing iminodiacetic acid groups. A concentrated solution of metals is obtained by eluting the resin with a small volume of 2 M nitric acid, which protonates the iminodiacetate groups. Chelex resin is often used for DNA extraction in preparation for polymerase chain reaction by binding to cations including Mg2+, which is an essential cofactor for DNase Deoxyribonuclease (DNase, for short) refers to a group of glycoprotein endonucleases which are enzymes that catalyze the hydrolytic cleavage of phosphodiester linkages in the DNA backbone, thus degrading DNA. The role of the DNase enzyme in cells ...s. Chelex protects the sample from DNases that might remain active after the boiling and could subsequently degrade the DNA, rendering it unsuitable for PCR. After boiling, the Che ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Extraction
The first isolation of deoxyribonucleic acid (DNA) was done in 1869 by Friedrich Miescher. Currently, it is a routine procedure in molecular biology or forensic analyses. For the chemical method, many different kits are used for extraction, and selecting the correct one will save time on kit optimization and extraction procedures. PCR sensitivity detection is considered to show the variation between the commercial kits. What does it deliver? DNA extraction is frequently a preliminary step in many diagnostic procedures used to identify environmental viruses and bacteria and diagnose illnesses and hereditary diseases. These methods consist of, but are not limited to: Fluorescence In Situ Hybridization (FISH) technique was developed in the 1980s. The basic idea is to use a nucleic acid probe to hybridize nuclear DNA from either interphase cells or metaphase chromosomes attached to a microscopic slide. It is a molecular method used, among other things, to recognize and count parti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
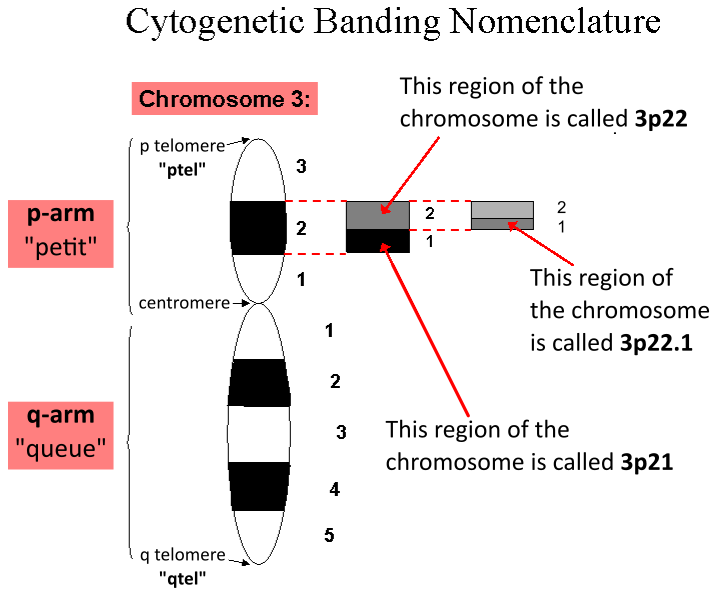
Locus (genetics)
In genetics, a locus (plural loci) is a specific, fixed position on a chromosome where a particular gene or genetic marker is located. Each chromosome carries many genes, with each gene occupying a different position or locus; in humans, the total number of protein-coding genes in a complete haploid set of 23 chromosomes is estimated at 19,000–20,000. Genes may possess multiple variants known as alleles, and an allele may also be said to reside at a particular locus. Diploid and polyploid cells whose chromosomes have the same allele at a given locus are called homozygous with respect to that locus, while those that have different alleles at a given locus are called heterozygous. The ordered list of loci known for a particular genome is called a gene map. Gene mapping is the process of determining the specific locus or loci responsible for producing a particular phenotype or biological trait. Association mapping, also known as "linkage disequilibrium mapping", is a method of ma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |