Machine learning in bioinformatics is the application of
machine learning
Machine learning (ML) is a field of study in artificial intelligence concerned with the development and study of Computational statistics, statistical algorithms that can learn from data and generalise to unseen data, and thus perform Task ( ...
algorithms to
bioinformatics
Bioinformatics () is an interdisciplinary field of science that develops methods and Bioinformatics software, software tools for understanding biological data, especially when the data sets are large and complex. Bioinformatics uses biology, ...
, including
genomics
Genomics is an interdisciplinary field of molecular biology focusing on the structure, function, evolution, mapping, and editing of genomes. A genome is an organism's complete set of DNA, including all of its genes as well as its hierarchical, ...
,
proteomics
Proteomics is the large-scale study of proteins. Proteins are vital macromolecules of all living organisms, with many functions such as the formation of structural fibers of muscle tissue, enzymatic digestion of food, or synthesis and replicatio ...
,
microarrays
A microarray is a multiplex lab-on-a-chip. Its purpose is to simultaneously detect the expression of thousands of biological interactions. It is a two-dimensional array on a solid substrateāusually a glass slide or silicon thin-film cellā ...
,
systems biology
Systems biology is the computational modeling, computational and mathematical analysis and modeling of complex biological systems. It is a biology-based interdisciplinary field of study that focuses on complex interactions within biological system ...
,
evolution
Evolution is the change in the heritable Phenotypic trait, characteristics of biological populations over successive generations. It occurs when evolutionary processes such as natural selection and genetic drift act on genetic variation, re ...
, and
text mining
Text mining, text data mining (TDM) or text analytics is the process of deriving high-quality information from text. It involves "the discovery by computer of new, previously unknown information, by automatically extracting information from differe ...
.
Prior to the emergence of machine learning, bioinformatics algorithms had to be programmed by hand; for problems such as
protein structure prediction
Protein structure prediction is the inference of the three-dimensional structure of a protein from its amino acid sequenceāthat is, the prediction of its Protein secondary structure, secondary and Protein tertiary structure, tertiary structure ...
, this proved difficult.
Machine learning techniques such as
deep learning
Deep learning is a subset of machine learning that focuses on utilizing multilayered neural networks to perform tasks such as classification, regression, and representation learning. The field takes inspiration from biological neuroscience a ...
can
learn features of data sets rather than requiring the programmer to define them individually. The algorithm can further learn how to combine low-level
features
Feature may refer to:
Computing
* Feature recognition, could be a hole, pocket, or notch
* Feature (computer vision), could be an edge, corner or blob
* Feature (machine learning), in statistics: individual measurable properties of the phenome ...
into more abstract features, and so on. This multi-layered approach allows such systems to make sophisticated predictions when appropriately trained. These methods contrast with other
computational biology
Computational biology refers to the use of techniques in computer science, data analysis, mathematical modeling and Computer simulation, computational simulations to understand biological systems and relationships. An intersection of computer sci ...
approaches which, while exploiting existing datasets, do not allow the data to be interpreted and analyzed in unanticipated ways.
Tasks
Machine learning algorithms in bioinformatics can be used for prediction, classification, and feature selection. Methods to achieve this task are varied and span many disciplines; most well known among them are machine learning and statistics. Classification and prediction tasks aim at building models that describe and distinguish classes or concepts for future prediction. The differences between them are the following:
* Classification/recognition outputs a categorical class, while prediction outputs a numerical valued feature.
* The type of algorithm, or process used to build the predictive models from data using analogies, rules, neural networks, probabilities, and/or statistics.
Due to the exponential growth of information technologies and applicable models, including artificial intelligence and data mining, in addition to the access ever-more comprehensive data sets, new and better information analysis techniques have been created, based on their ability to learn. Such models allow reach beyond description and provide insights in the form of testable models.
Approaches
Artificial neural networks
Artificial neural network
In machine learning, a neural network (also artificial neural network or neural net, abbreviated ANN or NN) is a computational model inspired by the structure and functions of biological neural networks.
A neural network consists of connected ...
s in bioinformatics have been used for:
* Comparing and aligning RNA, protein, and DNA sequences.
* Identification of promoters and finding genes from sequences related to DNA.
* Interpreting the expression-gene and micro-array data.
* Identifying the network (regulatory) of genes.
* Learning evolutionary relationships by constructing
phylogenetic tree
A phylogenetic tree or phylogeny is a graphical representation which shows the evolutionary history between a set of species or taxa during a specific time.Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA. In ...
s.
* Classifying and predicting
protein structure
Protein structure is the three-dimensional arrangement of atoms in an amino acid-chain molecule. Proteins are polymers specifically polypeptides formed from sequences of amino acids, which are the monomers of the polymer. A single amino acid ...
.
*
Molecular design and
docking
Feature engineering
The way that features, often vectors in a many-dimensional space, are extracted from the domain data is an important component of learning systems.
In genomics, a typical representation of a sequence is a vector of
k-mer
In bioinformatics, ''k''-mers are substrings of length k contained within a biological sequence. Primarily used within the context of computational genomics and sequence analysis, in which ''k''-mers are composed of nucleotides (''i.e''. A, T, ...
s frequencies, which is a vector of dimension
whose entries count the appearance of each subsequence of length
in a given sequence. Since for a value as small as
the dimensionality of these vectors is huge (e.g. in this case the dimension is
), techniques such as
principal component analysis
Principal component analysis (PCA) is a linear dimensionality reduction technique with applications in exploratory data analysis, visualization and data preprocessing.
The data is linearly transformed onto a new coordinate system such that th ...
are used to project the data to a lower dimensional space, thus selecting a smaller set of features from the sequences.
Classification
In this type of machine learning task, the output is a discrete variable. One example of this type of task in bioinformatics is labeling new genomic data (such as genomes of unculturable bacteria) based on a model of already labeled data.
Hidden Markov models
Hidden Markov models
A hidden Markov model (HMM) is a Markov model in which the observations are dependent on a latent (or ''hidden'') Markov process (referred to as X). An HMM requires that there be an observable process Y whose outcomes depend on the outcomes of X ...
(HMMs) are a class of
statistical models
A statistical model is a mathematical model that embodies a set of statistical assumptions concerning the generation of sample data (and similar data from a larger population). A statistical model represents, often in considerably idealized form ...
for sequential data (often related to systems evolving over time). An HMM is composed of two mathematical objects: an observed stateādependent process
, and an unobserved (hidden) state process
. In an HMM, the state process is not directly observed ā it is a 'hidden' (or 'latent') variable ā but observations are made of a stateādependent process (or observation process) that is driven by the underlying state process (and which can thus be regarded as a noisy measurement of the system states of interest). HMMs can be formulated in continuous time.
HMMs can be used to profile and convert a multiple sequence alignment into a position-specific scoring system suitable for searching databases for homologous sequences remotely. Additionally, ecological phenomena can be described by HMMs.
Convolutional neural networks
Convolutional neural network
A convolutional neural network (CNN) is a type of feedforward neural network that learns features via filter (or kernel) optimization. This type of deep learning network has been applied to process and make predictions from many different ty ...
s (CNN) are a class of
deep neural network
Deep learning is a subset of machine learning that focuses on utilizing multilayered neural network (machine learning), neural networks to perform tasks such as Statistical classification, classification, Regression analysis, regression, and re ...
whose architecture is based on shared weights of convolution kernels or filters that slide along input features, providing translation-equivariant responses known as feature maps.
CNNs take advantage of the hierarchical pattern in data and assemble patterns of increasing complexity using smaller and simpler patterns discovered via their filters.
Convolutional networks were
inspired by
biological
Biology is the scientific study of life and living organisms. It is a broad natural science that encompasses a wide range of fields and unifying principles that explain the structure, function, growth, origin, evolution, and distribution of ...
processes
in that the connectivity pattern between
neurons
A neuron (American English), neurone (British English), or nerve cell, is an membrane potential#Cell excitability, excitable cell (biology), cell that fires electric signals called action potentials across a neural network (biology), neural net ...
resembles the organization of the animal
visual cortex
The visual cortex of the brain is the area of the cerebral cortex that processes visual information. It is located in the occipital lobe. Sensory input originating from the eyes travels through the lateral geniculate nucleus in the thalam ...
. Individual
cortical neuron
The cerebral cortex, also known as the cerebral mantle, is the outer layer of neural tissue of the cerebrum of the brain in humans and other mammals. It is the largest site of neural integration in the central nervous system, and plays a key ...
s respond to stimuli only in a restricted region of the
visual field
The visual field is "that portion of space in which objects are visible at the same moment during steady fixation of the gaze in one direction"; in ophthalmology and neurology the emphasis is mostly on the structure inside the visual field and it i ...
known as the
receptive field
The receptive field, or sensory space, is a delimited medium where some physiological stimuli can evoke a sensory neuronal response in specific organisms.
Complexity of the receptive field ranges from the unidimensional chemical structure of od ...
. The receptive fields of different neurons partially overlap such that they cover the entire visual field.
CNN uses relatively little pre-processing compared to other
image classification algorithms. This means that the network learns to optimize the
filters
Filtration is a physical process that separates solid matter and fluid from a mixture.
Filter, filtering, filters or filtration may also refer to:
Science and technology
Computing
* Filter (higher-order function), in functional programming
* Fil ...
(or kernels) through automated learning, whereas in traditional algorithms these filters are
hand-engineered. This reduced reliance on prior knowledge of the analyst and on human intervention in manual feature extraction makes CNNs a desirable model.
A phylogenetic convolutional neural network (Ph-CNN) is a
convolutional neural network
A convolutional neural network (CNN) is a type of feedforward neural network that learns features via filter (or kernel) optimization. This type of deep learning network has been applied to process and make predictions from many different ty ...
architecture proposed by Fioranti et al. in 2018 to classify
metagenomics
Metagenomics is the study of all genetics, genetic material from all organisms in a particular environment, providing insights into their composition, diversity, and functional potential. Metagenomics has allowed researchers to profile the mic ...
data.
In this approach, phylogenetic data is endowed with patristic distance (the sum of the lengths of all branches connecting two
operational taxonomic unit
An operational taxonomic unit (OTU) is an operational definition used to classify groups of closely related individuals. The term was originally introduced in 1963 by Robert R. Sokal and Peter H. A. Sneath in the context of numerical taxonomy, wh ...
s
TU to select k-neighborhoods for each OTU, and each OTU and its neighbors are processed with convolutional filters.
Self-supervised learning
Unlike supervised methods,
self-supervised learning
Self-supervised learning (SSL) is a paradigm in machine learning where a model is trained on a task using the data itself to generate supervisory signals, rather than relying on externally-provided labels. In the context of neural networks, self ...
methods learn representations without relying on annotated data. That is well-suited for genomics, where
high throughput sequencing
DNA sequencing is the process of determining the nucleic acid sequence ā the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, thymine, cytosine, and guanine. The ...
techniques can create potentially large amounts of unlabeled data. Some examples of self-supervised learning methods applied on genomics include DNABERT and Self-GenomeNet.
Random forest
Random forest
Random forests or random decision forests is an ensemble learning method for statistical classification, classification, regression analysis, regression and other tasks that works by creating a multitude of decision tree learning, decision trees ...
s (RF) classify by constructing an ensemble of
decision trees
A decision tree is a decision support system, decision support recursive partitioning structure that uses a Tree (graph theory), tree-like Causal model, model of decisions and their possible consequences, including probability, chance event ou ...
, and outputting the average prediction of the individual trees. This is a modification of
bootstrap aggregating
Bootstrap aggregating, also called bagging (from bootstrap aggregating) or bootstrapping, is a machine learning (ML) ensemble meta-algorithm designed to improve the stability and accuracy of ML classification and regression algorithms. It also ...
(which aggregates a large collection of decision trees) and can be used for
classification
Classification is the activity of assigning objects to some pre-existing classes or categories. This is distinct from the task of establishing the classes themselves (for example through cluster analysis). Examples include diagnostic tests, identif ...
or
regression.
As random forests give an internal estimate of generalization error, cross-validation is unnecessary. In addition, they produce proximities, which can be used to impute missing values, and which enable novel data visualizations.
Computationally, random forests are appealing because they naturally handle both regression and (multiclass) classification, are relatively fast to train and to predict, depend only on one or two tuning parameters, have a built-in estimate of the generalization error, can be used directly for high-dimensional problems, and can easily be implemented in parallel. Statistically, random forests are appealing for additional features, such as measures of variable importance, differential class weighting, missing value imputation, visualization, outlier detection, and unsupervised learning.
Clustering
Clustering - the partitioning of a data set into disjoint subsets, so that the data in each subset are as close as possible to each other and as distant as possible from data in any other subset, according to some defined
distance
Distance is a numerical or occasionally qualitative measurement of how far apart objects, points, people, or ideas are. In physics or everyday usage, distance may refer to a physical length or an estimation based on other criteria (e.g. "two co ...
or
similarity function - is a common technique for statistical data analysis.
Clustering is central to much data-driven bioinformatics research and serves as a powerful computational method whereby means of hierarchical, centroid-based, distribution-based, density-based, and self-organizing maps classification, has long been studied and used in classical machine learning settings. Particularly, clustering helps to analyze unstructured and high-dimensional data in the form of sequences, expressions, texts, images, and so on. Clustering is also used to gain insights into biological processes at the
genomic
Genomics is an interdisciplinary field of molecular biology focusing on the structure, function, evolution, mapping, and editing of genomes. A genome is an organism's complete set of DNA, including all of its genes as well as its hierarchical, ...
level, e.g. gene functions, cellular processes, subtypes of cells,
gene regulation
Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products (protein or RNA). Sophisticated programs of gene expression are wide ...
, and metabolic processes.
Clustering algorithms used in bioinformatics
Data clustering algorithms can be hierarchical or partitional. Hierarchical algorithms find successive clusters using previously established clusters, whereas partitional algorithms determine all clusters at once. Hierarchical algorithms can be agglomerative (bottom-up) or divisive (top-down).
Agglomerative algorithms begin with each element as a separate cluster and merge them in successively larger clusters. Divisive algorithms begin with the whole set and proceed to divide it into successively smaller clusters.
Hierarchical clustering
In data mining and statistics, hierarchical clustering (also called hierarchical cluster analysis or HCA) is a method of cluster analysis that seeks to build a hierarchy of clusters. Strategies for hierarchical clustering generally fall into two ...
is calculated using metrics on
Euclidean spaces
Euclidean space is the fundamental space of geometry, intended to represent physical space. Originally, in Euclid's ''Elements'', it was the three-dimensional space of Euclidean geometry, but in modern mathematics there are ''Euclidean spaces'' ...
, the most commonly used is the
Euclidean distance
In mathematics, the Euclidean distance between two points in Euclidean space is the length of the line segment between them. It can be calculated from the Cartesian coordinates of the points using the Pythagorean theorem, and therefore is o ...
computed by finding the square of the difference between each variable, adding all the squares, and finding the square root of the said sum. An example of a
hierarchical clustering
In data mining and statistics, hierarchical clustering (also called hierarchical cluster analysis or HCA) is a method of cluster analysis that seeks to build a hierarchy of clusters. Strategies for hierarchical clustering generally fall into two ...
algorithm is
BIRCH
A birch is a thin-leaved deciduous hardwood tree of the genus ''Betula'' (), in the family Betulaceae, which also includes alders, hazels, and hornbeams. It is closely related to the beech- oak family Fagaceae. The genus ''Betula'' contains 3 ...
, which is particularly good on bioinformatics for its nearly linear
time complexity
In theoretical computer science, the time complexity is the computational complexity that describes the amount of computer time it takes to run an algorithm. Time complexity is commonly estimated by counting the number of elementary operations ...
given generally large datasets.
Partitioning algorithms are based on specifying an initial number of groups, and iteratively reallocating objects among groups to convergence. This algorithm typically determines all clusters at once. Most applications adopt one of two popular heuristic methods:
k-means
''k''-means clustering is a method of vector quantization, originally from signal processing, that aims to partition of a set, partition ''n'' observations into ''k'' clusters in which each observation belongs to the cluster (statistics), cluste ...
algorithm or
k-medoids
-medoids is a classical partitioning technique of clustering that splits the data set of objects into clusters, where the number of clusters assumed known ''a priori'' (which implies that the programmer must specify k before the execution of a - ...
. Other algorithms do not require an initial number of groups, such as
affinity propagation
In statistics and data mining, affinity propagation (AP) is a clustering algorithm based on the concept of "message passing" between data points.
Unlike clustering algorithms such as -means or -medoids, affinity propagation does not require the ...
. In a genomic setting this algorithm has been used both to cluster biosynthetic gene clusters in gene cluster families(GCF) and to cluster said GCFs.
Workflow
Typically, a workflow for applying machine learning to biological data goes through four steps:
* ''Recording,'' including capture and storage. In this step, different information sources may be merged into a single set.
* ''Preprocessing,'' including cleaning and restructuring into a ready-to-analyze form. In this step, uncorrected data are eliminated or corrected, while missing data maybe imputed and relevant variables chosen.
* ''Analysis,'' evaluating data using either supervised or unsupervised algorithms. The algorithm is typically trained on a subset of data, optimizing parameters, and evaluated on a separate test subset.
* ''Visualization and interpretation,'' where knowledge is represented effectively using different methods to assess the significance and importance of the findings.
Data errors
* Duplicate data is a significant issue in bioinformatics. Publicly available data may be of uncertain quality.
* Errors during experimentation.
* Erroneous interpretation.
* Typing mistakes.
* Non-standardized methods (3D structure in PDB from multiple sources, X-ray diffraction, theoretical modeling, nuclear magnetic resonance, etc.) are used in experiments.
Applications
In general, a machine learning system can usually be trained to recognize elements of a certain class given sufficient samples.
For example, machine learning methods can be trained to identify specific visual features such as splice sites.
Support vector machines
In machine learning, support vector machines (SVMs, also support vector networks) are supervised max-margin models with associated learning algorithms that analyze data for classification and regression analysis. Developed at AT&T Bell Laborato ...
have been extensively used in cancer genomic studies.
In addition,
deep learning
Deep learning is a subset of machine learning that focuses on utilizing multilayered neural networks to perform tasks such as classification, regression, and representation learning. The field takes inspiration from biological neuroscience a ...
has been incorporated into bioinformatic algorithms. Deep learning applications have been used for regulatory genomics and cellular imaging.
Other applications include medical image classification, genomic sequence analysis, as well as protein structure classification and prediction.
Deep learning has been applied to regulatory genomics, variant calling and pathogenicity scores.
Natural language processing
Natural language processing (NLP) is a subfield of computer science and especially artificial intelligence. It is primarily concerned with providing computers with the ability to process data encoded in natural language and is thus closely related ...
and
text mining
Text mining, text data mining (TDM) or text analytics is the process of deriving high-quality information from text. It involves "the discovery by computer of new, previously unknown information, by automatically extracting information from differe ...
have helped to understand phenomena including protein-protein interaction, gene-disease relation as well as predicting biomolecule structures and functions.
Precision/personalized medicine
Natural language processing
Natural language processing (NLP) is a subfield of computer science and especially artificial intelligence. It is primarily concerned with providing computers with the ability to process data encoded in natural language and is thus closely related ...
algorithms
personalized medicine for patients who suffer genetic diseases, by combining the extraction of clinical information and genomic data available from the patients. Institutes such as Health-funded Pharmacogenomics Research Network focus on finding breast cancer treatments.
Precision medicine
Precision, precise or precisely may refer to:
Arts and media
* ''Precision'' (march), the official marching music of the Royal Military College of Canada
* "Precision" (song), by Big Sean
* ''Precisely'' (sketch), a dramatic sketch by the Eng ...
considers individual genomic variability, enabled by large-scale biological databases. Machine learning can be applied to perform the matching function between (groups of patients) and specific treatment modalities.
Computational techniques are used to solve other problems, such as efficient primer design for
PCR, biological-image analysis and back translation of proteins (which is, given the degeneration of the genetic code, a complex combinatorial problem).
Genomics

While genomic sequence data has historically been sparse due to the technical difficulty of sequencing a piece of DNA, the number of available sequences is growing. On average, the number of
bases available in the
GenBank
The GenBank sequence database is an open access, annotated collection of all publicly available nucleotide sequences and their protein translations. It is produced and maintained by the National Center for Biotechnology Information (NCBI; a par ...
public repository has doubled every 18 months since 1982. However, while
raw data
Raw data, also known as primary data, are ''data'' (e.g., numbers, instrument readings, figures, etc.) collected from a source. In the context of examinations, the raw data might be described as a raw score (after test scores).
If a scientist ...
was becoming increasingly available and accessible, , biological interpretation of this data was occurring at a much slower pace.
This made for an increasing need for developing
computational genomics Computational genomics refers to the use of computational and statistical analysis to decipher biology from genome sequences and related data, including both DNA and RNA sequence as well as other "post-genomic" data (i.e., experimental data obtained ...
tools, including machine learning systems, that can automatically determine the location of protein-encoding genes within a given DNA sequence (i.e.
gene prediction
In computational biology, gene prediction or gene finding refers to the process of identifying the regions of genomic DNA that encode genes. This includes protein-coding genes as well as RNA genes, but may also include prediction of other functio ...
).
Gene prediction is commonly performed through both ''extrinsic searches'' and ''intrinsic searches''.
For the extrinsic search, the input DNA sequence is run through a large database of sequences whose genes have been previously discovered and their locations annotated and identifying the target sequence's genes by determining which strings of bases within the sequence are
homologous to known gene sequences. However, not all the genes in a given input sequence can be identified through homology alone, due to limits in the size of the database of known and annotated gene sequences. Therefore, an intrinsic search is needed where a gene prediction program attempts to identify the remaining genes from the DNA sequence alone.
Machine learning has also been used for the problem of
multiple sequence alignment
Multiple sequence alignment (MSA) is the process or the result of sequence alignment of three or more biological sequences, generally protein, DNA, or RNA. These alignments are used to infer evolutionary relationships via phylogenetic analysis an ...
which involves aligning many DNA or amino acid sequences in order to determine regions of similarity that could indicate a shared evolutionary history.
It can also be used to detect and visualize genome rearrangements.
Proteomics

Protein
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residue (biochemistry), residues. Proteins perform a vast array of functions within organisms, including Enzyme catalysis, catalysing metab ...
s, strings of
amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although over 500 amino acids exist in nature, by far the most important are the 22 Ī±-amino acids incorporated into proteins. Only these 22 a ...
s, gain much of their function from
protein folding
Protein folding is the physical process by which a protein, after Protein biosynthesis, synthesis by a ribosome as a linear chain of Amino acid, amino acids, changes from an unstable random coil into a more ordered protein tertiary structure, t ...
, where they conform into a three-dimensional structure, including the
primary structure
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthe ...
, the
secondary structure
Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta ...
(
alpha helices
An alpha helix (or Ī±-helix) is a sequence of amino acids in a protein that are twisted into a coil (a helix).
The alpha helix is the most common structural arrangement in the secondary structure of proteins. It is also the most extreme type of l ...
and
beta sheet
The beta sheet (Ī²-sheet, also Ī²-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (Ī²-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a gene ...
s), the
tertiary structure
Protein tertiary structure is the three-dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains and the ...
, and the
quaternary structure
Protein quaternary structure is the fourth (and highest) classification level of protein structure. Protein quaternary structure refers to the structure of proteins which are themselves composed of two or more smaller protein chains (also refe ...
.
Protein secondary structure prediction is a main focus of this subfield as tertiary and quaternary structures are determined based on the secondary structure.
Solving the true structure of a protein is expensive and time-intensive, furthering the need for systems that can accurately predict the structure of a protein by analyzing the amino acid sequence directly.
Prior to machine learning, researchers needed to conduct this prediction manually. This trend began in 1951 when Pauling and Corey released their work on predicting the hydrogen bond configurations of a protein from a polypeptide chain. Automatic feature learning reaches an accuracy of 82-84%.
Recent approaches have utilized deep learning techniques for state-of-the-art secondary structure predictions. For example, DeepCNF (deep convolutional neural fields) achieved an accuracy of approximately 84% when tasked to classify the amino acids of a protein sequence into one of three structural classes (helix, sheet, or coil).
The theoretical limit for three-state protein secondary structure is 88ā90%.
In 2018,
AlphaFold
AlphaFold is an artificial intelligence (AI) program developed by DeepMind, a subsidiary of Alphabet, which performs predictions of protein structure. It is designed using deep learning techniques.
AlphaFold 1 (2018) placed first in the overall ...
, an
artificial intelligence
Artificial intelligence (AI) is the capability of computer, computational systems to perform tasks typically associated with human intelligence, such as learning, reasoning, problem-solving, perception, and decision-making. It is a field of re ...
(AI) program developed by
DeepMind
DeepMind Technologies Limited, trading as Google DeepMind or simply DeepMind, is a BritishāAmerican artificial intelligence research laboratory which serves as a subsidiary of Alphabet Inc. Founded in the UK in 2010, it was acquired by Go ...
, placed first in the overall rankings of the 13th
Critical Assessment of Structure Prediction (CASP). It was particularly successful at predicting the most accurate structures for targets rated as most difficult by the competition organizers, where no existing
template structures were available from
proteins
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, re ...
with partially similar sequences. AlphaFold 2 (2020) repeated this placement in the CASP14 competition and achieved a level of accuracy much higher than any other entry.
Machine learning has also been applied to proteomics problems such as
protein side-chain prediction,
protein loop modeling, and
protein contact map
A protein contact map represents the distance between all possible Amino acid, amino acid residue pairs of a three-dimensional protein structure using a binary two-dimensional Matrix (mathematics), matrix. For two residues i and j, the ij element o ...
prediction.
Metagenomics
Metagenomics
Metagenomics is the study of all genetics, genetic material from all organisms in a particular environment, providing insights into their composition, diversity, and functional potential. Metagenomics has allowed researchers to profile the mic ...
is the study of microbial communities from environmental DNA samples. Currently, limitations and challenges predominate in the implementation of machine learning tools due to the amount of data in environmental samples. Supercomputers and web servers have made access to these tools easier.
The high dimensionality of microbiome datasets is a major challenge in studying the microbiome; this significantly limits the power of current approaches for identifying true differences and increases the chance of false discoveries.
Despite their importance, machine learning tools related to metagenomics have focused on the study of gut microbiota and the relationship with digestive diseases, such as
inflammatory bowel disease
Inflammatory bowel disease (IBD) is a group of inflammatory conditions of the colon and small intestine, with Crohn's disease and ulcerative colitis (UC) being the principal types. Crohn's disease affects the small intestine and large intestine ...
(IBD), ''
Clostridioides difficile
''Clostridioides difficile'' ( syn. ''Clostridium difficile'') is a bacterium known for causing serious diarrheal infections, and may also cause colon cancer. It is known also as ''C. difficile'', or ''C. diff'' (), and is a Gram-positive spec ...
'' infection (CDI),
colorectal cancer
Colorectal cancer (CRC), also known as bowel cancer, colon cancer, or rectal cancer, is the development of cancer from the Colon (anatomy), colon or rectum (parts of the large intestine). Signs and symptoms may include Lower gastrointestinal ...
and
diabetes
Diabetes mellitus, commonly known as diabetes, is a group of common endocrine diseases characterized by sustained high blood sugar levels. Diabetes is due to either the pancreas not producing enough of the hormone insulin, or the cells of th ...
, seeking better diagnosis and treatments.
Many algorithms were developed to classify microbial communities according to the health condition of the host, regardless of the type of sequence data, e.g.
16S rRNA
16S ribosomal RNA (or 16Svedberg, S rRNA) is the RNA component of the 30S subunit of a prokaryotic ribosome (SSU rRNA). It binds to the Shine-Dalgarno sequence and provides most of the SSU structure.
The genes coding for it are referred to as ...
or
whole-genome sequencing
Whole genome sequencing (WGS), also known as full genome sequencing or just genome sequencing, is the process of determining the entirety of the DNA sequence of an organism's genome at a single time. This entails sequencing all of an organism's ...
(WGS), using methods such as least absolute shrinkage and selection operator classifier,
random forest
Random forests or random decision forests is an ensemble learning method for statistical classification, classification, regression analysis, regression and other tasks that works by creating a multitude of decision tree learning, decision trees ...
, supervised classification model, and gradient boosted tree model.
Neural networks
A neural network is a group of interconnected units called neurons that send signals to one another. Neurons can be either Cell (biology), biological cells or signal pathways. While individual neurons are simple, many of them together in a netwo ...
, such as
recurrent neural network
Recurrent neural networks (RNNs) are a class of artificial neural networks designed for processing sequential data, such as text, speech, and time series, where the order of elements is important. Unlike feedforward neural networks, which proces ...
s (RNN),
convolutional neural network
A convolutional neural network (CNN) is a type of feedforward neural network that learns features via filter (or kernel) optimization. This type of deep learning network has been applied to process and make predictions from many different ty ...
s (CNN), and
Hopfield neural networks have been added.
For example, in 2018, Fioravanti et al. developed an algorithm called Ph-CNN to classify data samples from healthy patients and patients with IBD symptoms (to distinguish healthy and sick patients) by using phylogenetic trees and convolutional neural networks.
In addition,
random forest
Random forests or random decision forests is an ensemble learning method for statistical classification, classification, regression analysis, regression and other tasks that works by creating a multitude of decision tree learning, decision trees ...
(RF) methods and implemented importance measures help in the identification of microbiome species that can be used to distinguish diseased and non-diseased samples. However, the performance of a decision tree and the diversity of decision trees in the ensemble significantly influence the performance of RF algorithms. The generalization error for RF measures how accurate the individual classifiers are and their interdependence. Therefore, the high dimensionality problems of microbiome datasets pose challenges. Effective approaches require many possible variable combinations, which exponentially increases the computational burden as the number of features increases.
For microbiome analysis in 2020 Dang & Kishino
developed a novel analysis pipeline. The core of the pipeline is an RF classifier coupled with forwarding
variable selection (RF-FVS), which selects a minimum-size core set of microbial species or functional signatures that maximize the predictive classifier performance. The framework combines:
* identifying a few significant features by a massively parallel forward
variable selection procedure
* mapping the selected species on a
phylogenetic tree
A phylogenetic tree or phylogeny is a graphical representation which shows the evolutionary history between a set of species or taxa during a specific time.Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA. In ...
, and
* predicting functional profiles by functional gene enrichment analysis from metagenomic
16S rRNA
16S ribosomal RNA (or 16Svedberg, S rRNA) is the RNA component of the 30S subunit of a prokaryotic ribosome (SSU rRNA). It binds to the Shine-Dalgarno sequence and provides most of the SSU structure.
The genes coding for it are referred to as ...
data.
They demonstrated performance by analyzing two published datasets from large-scale case-control studies:
* 16S rRNA gene amplicon data for ''C. difficile'' infection (CDI) and
* shotgun metagenomics data for human colorectal cancer (CRC).
The proposed approach improved the accuracy from 81% to 99.01% for CDI and from 75.14% to 90.17% for CRC.
The use of machine learning in environmental samples has been less explored, maybe because of data complexity, especially from WGS. Some works show that it is possible to apply these tools in environmental samples. In 2021 Dhungel et al., designed an R package called MegaR. This package allows working with 16S rRNA and whole metagenomic sequences to make taxonomic profiles and classification models by machine learning models. MegaR includes a comfortable visualization environment to improve the user experience. Machine learning in environmental metagenomics can help to answer questions related to the interactions between microbial communities and ecosystems, e.g. the work of Xun et al., in 2021 where the use of different machine learning methods offered insights on the relationship among the soil, microbiome biodiversity, and ecosystem stability.
Microarrays
Microarray
A microarray is a multiplex (assay), multiplex lab-on-a-chip. Its purpose is to simultaneously detect the expression of thousands of biological interactions. It is a two-dimensional array on a Substrate (materials science), solid substrateāusu ...
s, a type of
lab-on-a-chip
A lab-on-a-chip (LOC) is a device that integrates one or several laboratory functions on a single integrated circuit (commonly called a "chip") of only millimeters to a few square centimeters to achieve automation and high-throughput screening. ...
, are used for automatically collecting data about large amounts of biological material. Machine learning can aid in analysis, and has been applied to expression pattern identification, classification, and genetic network induction.
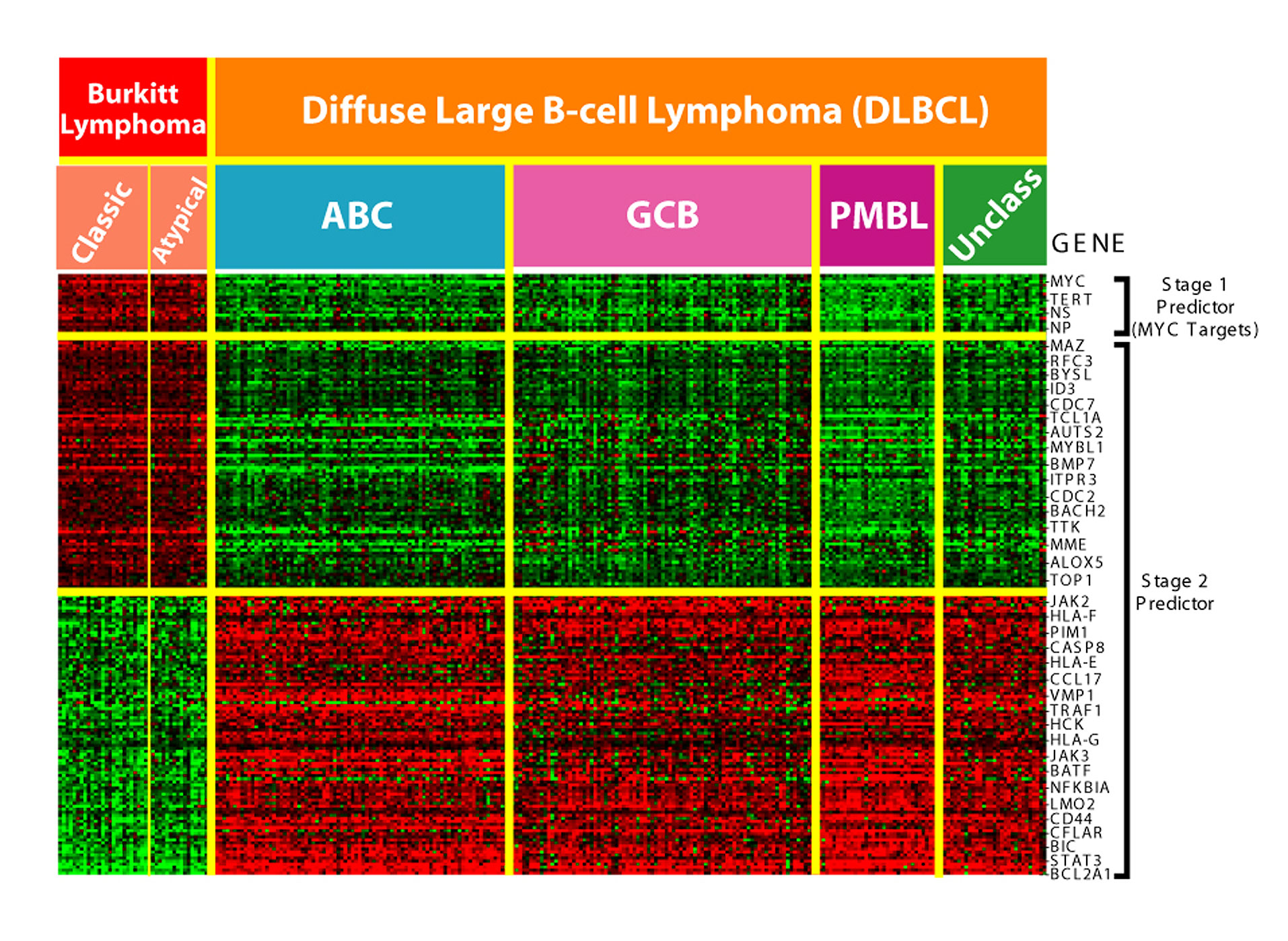
This technology is especially useful for monitoring gene expression, aiding in diagnosing cancer by examining which genes are expressed.
One of the main tasks is identifying which genes are expressed based on the collected data.
In addition, due to the huge number of genes on which data is collected by the microarray, winnowing the large amount of irrelevant data to the task of expressed gene identification is challenging. Machine learning presents a potential solution as various classification methods can be used to perform this identification. The most commonly used methods are
radial basis function network
In the field of mathematical modeling, a radial basis function network is an artificial neural network that uses radial basis functions as activation functions. The output of the network is a linear combination of radial basis functions of the in ...
s,
deep learning
Deep learning is a subset of machine learning that focuses on utilizing multilayered neural networks to perform tasks such as classification, regression, and representation learning. The field takes inspiration from biological neuroscience a ...
,
Bayesian classification,
decision tree
A decision tree is a decision support system, decision support recursive partitioning structure that uses a Tree (graph theory), tree-like Causal model, model of decisions and their possible consequences, including probability, chance event ou ...
s, and
random forest
Random forests or random decision forests is an ensemble learning method for statistical classification, classification, regression analysis, regression and other tasks that works by creating a multitude of decision tree learning, decision trees ...
.
Systems biology
Systems biology focuses on the study of emergent behaviors from complex interactions of simple biological components in a system. Such components can include DNA, RNA, proteins, and metabolites.
Machine learning has been used to aid in modeling these interactions in domains such as genetic networks, signal transduction networks, and metabolic pathways.
, a machine learning technique for determining the relationship between different variables, are one of the most commonly used methods for modeling genetic networks.
In addition, machine learning has been applied to systems biology problems such as identifying
transcription factor binding sites using
Markov chain optimization.
Genetic algorithm
In computer science and operations research, a genetic algorithm (GA) is a metaheuristic inspired by the process of natural selection that belongs to the larger class of evolutionary algorithms (EA). Genetic algorithms are commonly used to g ...
s, machine learning techniques which are based on the natural process of evolution, have been used to model genetic networks and regulatory structures.
Other systems biology applications of machine learning include the task of enzyme function prediction, high throughput microarray data analysis, analysis of genome-wide association studies to better understand markers of disease, protein function prediction.
Evolution
This domain, particularly
phylogenetic tree
A phylogenetic tree or phylogeny is a graphical representation which shows the evolutionary history between a set of species or taxa during a specific time.Felsenstein J. (2004). ''Inferring Phylogenies'' Sinauer Associates: Sunderland, MA. In ...
reconstruction, uses the features of machine learning techniques. Phylogenetic trees are schematic representations of the evolution of organisms. Initially, they were constructed using features such as morphological and metabolic features. Later, due to the availability of genome sequences, the construction of the phylogenetic tree algorithm used the concept based on genome comparison. With the help of optimization techniques, a comparison was done by means of multiple sequence alignment.
Stroke diagnosis
Machine learning methods for the analysis of
neuroimaging
Neuroimaging is the use of quantitative (computational) techniques to study the neuroanatomy, structure and function of the central nervous system, developed as an objective way of scientifically studying the healthy human brain in a non-invasive ...
data are used to help diagnose
stroke
Stroke is a medical condition in which poor cerebral circulation, blood flow to a part of the brain causes cell death. There are two main types of stroke: brain ischemia, ischemic, due to lack of blood flow, and intracranial hemorrhage, hemor ...
. Historically multiple approaches to this problem involved neural networks.
Multiple approaches to detect strokes used machine learning. As proposed by Mirtskhulava, feed-forward networks were tested to detect strokes using neural imaging. As proposed by Titano 3D-CNN techniques were tested in supervised classification to screen head CT images for acute neurologic events. Three-dimensional
CNN
Cable News Network (CNN) is a multinational news organization operating, most notably, a website and a TV channel headquartered in Atlanta. Founded in 1980 by American media proprietor Ted Turner and Reese Schonfeld as a 24-hour cable ne ...
and
SVM methods are often used.
Text mining
The increase in biological publications increased the difficulty in searching and compiling relevant available information on a given topic. This task is known as
knowledge extraction
Knowledge extraction is the creation of knowledge from structured ( relational databases, XML) and unstructured (text, documents, images) sources. The resulting knowledge needs to be in a machine-readable and machine-interpretable format and must ...
. It is necessary for biological data collection which can then in turn be fed into machine learning algorithms to generate new biological knowledge.
Machine learning can be used for this knowledge extraction task using techniques such as
natural language processing
Natural language processing (NLP) is a subfield of computer science and especially artificial intelligence. It is primarily concerned with providing computers with the ability to process data encoded in natural language and is thus closely related ...
to extract the useful information from human-generated reports in a database.
Text Nailing, an alternative approach to machine learning, capable of extracting features from clinical narrative notes was introduced in 2017.
This technique has been applied to the search for novel drug targets, as this task requires the examination of information stored in biological databases and journals.
Annotations of proteins in protein databases often do not reflect the complete known set of knowledge of each protein, so additional information must be extracted from biomedical literature. Machine learning has been applied to the automatic annotation of gene and protein function, determination of the
protein subcellular localization,
DNA-expression array analysis, large-scale
protein interaction
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, respon ...
analysis, and molecule interaction analysis.
Another application of text mining is the detection and visualization of distinct DNA regions given sufficient reference data.
Clustering and abundance profiling of biosynthetic gene clusters
Microbial communities are complex assembles of diverse microorganisms, where symbiont partners constantly produce diverse metabolites derived from the primary and secondary (specialized) metabolism, from which metabolism plays an important role in microbial interaction. Metagenomic and metatranscriptomic data are an important source for deciphering communications signals.
Molecular mechanisms produce specialized metabolites in various ways.
Biosynthetic Gene Clusters (BGCs) attract attention, since several metabolites are clinically valuable, anti-microbial, anti-fungal, anti-parasitic, anti-tumor and immunosuppressive agents produced by the modular action of multi-enzymatic, multi-domains gene clusters, such as
Nonribosomal peptide
Nonribosomal peptides (NRP) are a class of peptide secondary metabolites, usually produced by microorganisms like bacterium, bacteria and fungi. Nonribosomal peptides are also found in higher organisms, such as nudibranchs, but are thought to be ma ...
synthetases (NRPSs) and
polyketide synthase
Polyketide synthases (PKSs) are a family of multi- domain enzymes or enzyme complexes that produce polyketides, a large class of secondary metabolites, in bacteria, fungi, plants, and a few animal lineages. The biosyntheses of polyketides share ...
s (PKSs). Diverse studies
show that grouping BGCs that share homologous core genes into gene cluster families (GCFs) can yield useful insights into the chemical diversity of the analyzed strains, and can support linking BGCs to their secondary metabolites.
GCFs have been used as functional markers in human health studies and to study the ability of soil to suppress fungal pathogens. Given their direct relationship to catalytic enzymes, and compounds produced from their encoded pathways, BGCs/GCFs can serve as a proxy to explore the chemical space of microbial secondary metabolism. Cataloging GCFs in sequenced microbial genomes yields an overview of the existing chemical diversity and offers insights into future priorities.
Tools such as BiG-SLiCE and BIG-MAP
have emerged with the sole purpose of unveiling the importance of BGCs in natural environments.
Decodification of RiPPs chemical structures
The increase of experimentally characterized
(RiPPs), together with the availability of information on their sequence and chemical structure, selected from databases such as BAGEL, BACTIBASE, MIBIG, and THIOBASE, provide the opportunity to develop machine learning tools to decode the chemical structure and classify them.
In 2017, researchers at the National Institute of Immunology of New Delhi, India, developed RiPPMiner software, a bioinformatics resource for decoding RiPP chemical structures by genome mining. The RiPPMiner web server consists of a query interface and the RiPPDB database. RiPPMiner defines 12 subclasses of RiPPs, predicting the cleavage site of the leader peptide and the final cross-link of the RiPP chemical structure.
Mass spectral similarity scoring
Many tandem mass spectrometry (MS/MS) based metabolomics studies, such as library matching and molecular networking, use spectral similarity as a proxy for structural similarity. Spec2vec algorithm provides a new way of spectral similarity score, based on
Word2Vec
Word2vec is a technique in natural language processing (NLP) for obtaining vector representations of words. These vectors capture information about the meaning of the word based on the surrounding words. The word2vec algorithm estimates these rep ...
. Spec2Vec learns fragmental relationships within a large set of spectral data, in order to assess spectral similarities between molecules and to classify unknown molecules through these comparisons.
For systemic annotation, some metabolomics studies rely on fitting measured fragmentation mass spectra to library spectra or contrasting spectra via network analysis. Scoring functions are used to determine the similarity between pairs of fragment spectra as part of these processes. So far, no research has suggested scores that are significantly different from the commonly utilized
cosine-based similarity.
Databases
An important part of bioinformatics is the management of big datasets, known as databases of reference. Databases exist for each type of biological data, for example for biosynthetic gene clusters and metagenomes.
General databases by bioinformatics
National Center for Biotechnology Information
The National Center for Biotechnology Information (NCBI) provides a large suite of online resources for biological information and data, including the
GenBank
The GenBank sequence database is an open access, annotated collection of all publicly available nucleotide sequences and their protein translations. It is produced and maintained by the National Center for Biotechnology Information (NCBI; a par ...
nucleic acid sequence database and the
PubMed
PubMed is an openly accessible, free database which includes primarily the MEDLINE database of references and abstracts on life sciences and biomedical topics. The United States National Library of Medicine (NLM) at the National Institute ...
database of citations and abstracts for published life science journals. Augmenting many of the Web applications are custom implementations of the BLAST program optimized to search specialized data sets. Resources include PubMed Data Management, RefSeq Functional Elements, genome data download, variation services API, Magic-BLAST, QuickBLASTp, and Identical Protein Groups. All of these resources can be accessed through NCBI.
Bioinformatics analysis for biosynthetic gene clusters
antiSMASH
antiSMASH allows the rapid genome-wide identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genomes. It integrates and cross-links with a large number of in silico
secondary metabolite
Secondary metabolites, also called ''specialised metabolites'', ''secondary products'', or ''natural products'', are organic compounds produced by any lifeform, e.g. bacteria, archaea, fungi, animals, or plants, which are not directly involved ...
analysis tools.
gutSMASH
gutSMASH is a tool that systematically evaluates bacterial metabolic potential by predicting both known and novel
anaerobic
Anaerobic means "living, active, occurring, or existing in the absence of free oxygen", as opposed to aerobic which means "living, active, or occurring only in the presence of oxygen." Anaerobic may also refer to:
*Adhesive#Anaerobic, Anaerobic ad ...
metabolic gene clusters (MGCs) from the gut
microbiome
A microbiome () is the community of microorganisms that can usually be found living together in any given habitat. It was defined more precisely in 1988 by Whipps ''et al.'' as "a characteristic microbial community occupying a reasonably wel ...
.
MIBiG
MIBiG, the minimum information about a biosynthetic gene cluster specification, provides a standard for annotations and
metadata
Metadata (or metainformation) is "data that provides information about other data", but not the content of the data itself, such as the text of a message or the image itself. There are many distinct types of metadata, including:
* Descriptive ...
on biosynthetic gene clusters and their molecular products. MIBiG is a Genomic Standards Consortium project that builds on the minimum information about any sequence (MIxS) framework.
MIBiG facilitates the standardized deposition and retrieval of biosynthetic gene cluster data as well as the development of comprehensive comparative analysis tools. It empowers next-generation research on the biosynthesis, chemistry and ecology of broad classes of societally relevant bioactive
secondary metabolites
Secondary metabolites, also called ''specialised metabolites'', ''secondary products'', or ''natural products'', are organic compounds produced by any lifeform, e.g. bacteria, archaea, fungi, animals, or plants, which are not directly involved ...
, guided by robust experimental evidence and rich metadata components.
SILVA
SILVA is an interdisciplinary project among biologists and computers scientists assembling a complete database of RNA ribosomal (rRNA) sequences of genes, both small (
16S,
18S, SSU) and large (
23S,
28S, LSU) subunits, which belong to the bacteria, archaea and eukarya domains. These data are freely available for academic and commercial use.
Greengenes
Greengenes is a full-length
16S rRNA
16S ribosomal RNA (or 16Svedberg, S rRNA) is the RNA component of the 30S subunit of a prokaryotic ribosome (SSU rRNA). It binds to the Shine-Dalgarno sequence and provides most of the SSU structure.
The genes coding for it are referred to as ...
gene database that provides chimera screening, standard alignment and a curated taxonomy based on de novo tree inference. Overview:
* 1,012,863 RNA sequences from 92,684 organisms contributed to RNAcentral.
* The shortest sequence has 1,253 nucleotides, the longest 2,368.
* The average length is 1,402 nucleotides.
* Database version: 13.5.
Open Tree of Life Taxonomy
Open Tree of Life Taxonomy (OTT) aims to build a complete, dynamic, and digitally available Tree of Life by synthesizing published phylogenetic trees along with taxonomic data. Phylogenetic trees have been classified, aligned, and merged. Taxonomies have been used to fill in sparse regions and gaps left by phylogenies. OTT is a base that has been little used for sequencing analyzes of the 16S region, however, it has a greater number of sequences classified taxonomically down to the genus level compared to SILVA and Greengenes. However, in terms of classification at the edge level, it contains a lesser amount of information
Ribosomal Database Project
Ribosomal Database Project (RDP) is a database that provides RNA ribosomal (rRNA) sequences of small subunits of domain bacterial and archaeal (
16S); and fungal rRNA sequences of large subunits (
28S).
References
__FORCETOC__
{{Differentiable computing
Machine learning
Bioinformatics
 While genomic sequence data has historically been sparse due to the technical difficulty of sequencing a piece of DNA, the number of available sequences is growing. On average, the number of bases available in the
While genomic sequence data has historically been sparse due to the technical difficulty of sequencing a piece of DNA, the number of available sequences is growing. On average, the number of bases available in the 
 This technology is especially useful for monitoring gene expression, aiding in diagnosing cancer by examining which genes are expressed. One of the main tasks is identifying which genes are expressed based on the collected data. In addition, due to the huge number of genes on which data is collected by the microarray, winnowing the large amount of irrelevant data to the task of expressed gene identification is challenging. Machine learning presents a potential solution as various classification methods can be used to perform this identification. The most commonly used methods are
This technology is especially useful for monitoring gene expression, aiding in diagnosing cancer by examining which genes are expressed. One of the main tasks is identifying which genes are expressed based on the collected data. In addition, due to the huge number of genes on which data is collected by the microarray, winnowing the large amount of irrelevant data to the task of expressed gene identification is challenging. Machine learning presents a potential solution as various classification methods can be used to perform this identification. The most commonly used methods are