|
Affinity Propagation
In statistics and data mining, affinity propagation (AP) is a Cluster analysis, clustering algorithm based on the concept of "message passing" between data points. Unlike clustering algorithms such as K-means clustering, -means or K-medoids, -medoids, affinity propagation does not require the number of clusters to be determined or estimated before running the algorithm. Similar to -medoids, affinity propagation finds "exemplars," members of the input set that are representative of clusters. Algorithm Let through be a set of data points, with no assumptions made about their internal structure, and let be a function that quantifies the similarity between any two points, such that if and only if, iff is more similar to than to . For this example, the negative squared distance of two data points was used i.e. for points and , s(i,k) = - \left\, x_i - x_k \right\, ^2 The diagonal of (i.e. s(i,i)) is particularly important, as it represents the instance preference, meaning how l ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Statistics
Statistics (from German language, German: ''wikt:Statistik#German, Statistik'', "description of a State (polity), state, a country") is the discipline that concerns the collection, organization, analysis, interpretation, and presentation of data. In applying statistics to a scientific, industrial, or social problem, it is conventional to begin with a statistical population or a statistical model to be studied. Populations can be diverse groups of people or objects such as "all people living in a country" or "every atom composing a crystal". Statistics deals with every aspect of data, including the planning of data collection in terms of the design of statistical survey, surveys and experimental design, experiments.Dodge, Y. (2006) ''The Oxford Dictionary of Statistical Terms'', Oxford University Press. When census data cannot be collected, statisticians collect data by developing specific experiment designs and survey sample (statistics), samples. Representative sampling as ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Markov Clustering
In statistics, Markov chain Monte Carlo (MCMC) methods comprise a class of algorithms for sampling from a probability distribution. By constructing a Markov chain that has the desired distribution as its equilibrium distribution, one can obtain a sample of the desired distribution by recording states from the chain. The more steps that are included, the more closely the distribution of the sample matches the actual desired distribution. Various algorithms exist for constructing chains, including the Metropolis–Hastings algorithm. Application domains MCMC methods are primarily used for calculating numerical approximations of multi-dimensional integrals, for example in Bayesian statistics, computational physics, computational biology and computational linguistics. In Bayesian statistics, the recent development of MCMC methods has made it possible to compute large hierarchical models that require integrations over hundreds to thousands of unknown parameters. In rare event sa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Scikit-learn
scikit-learn (formerly scikits.learn and also known as sklearn) is a free software machine learning library for the Python programming language. It features various classification, regression and clustering algorithms including support-vector machines, random forests, gradient boosting, ''k''-means and DBSCAN, and is designed to interoperate with the Python numerical and scientific libraries NumPy and SciPy. Scikit-learn is a NumFOCUS fiscally sponsored project. Overview The scikit-learn project started as scikits.learn, a Google Summer of Code project by French data scientist David Cournapeau. The name of the project stems from the notion that it is a "SciKit" (SciPy Toolkit), a separately developed and distributed third-party extension to SciPy. The original codebase was later rewritten by other developers. In 2010, contributors Fabian Pedregosa, Gaël Varoquaux, Alexandre Gramfort and Vincent Michel, from the French Institute for Research in Computer Science and Automation ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Python (programming Language)
Python is a high-level, general-purpose programming language. Its design philosophy emphasizes code readability with the use of significant indentation. Python is dynamically-typed and garbage-collected. It supports multiple programming paradigms, including structured (particularly procedural), object-oriented and functional programming. It is often described as a "batteries included" language due to its comprehensive standard library. Guido van Rossum began working on Python in the late 1980s as a successor to the ABC programming language and first released it in 1991 as Python 0.9.0. Python 2.0 was released in 2000 and introduced new features such as list comprehensions, cycle-detecting garbage collection, reference counting, and Unicode support. Python 3.0, released in 2008, was a major revision that is not completely backward-compatible with earlier versions. Python 2 was discontinued with version 2.7.18 in 2020. Python consistently ranks as ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Julia (programming Language)
Julia is a high-level, dynamic programming language. Its features are well suited for numerical analysis and computational science. Distinctive aspects of Julia's design include a type system with parametric polymorphism in a dynamic programming language; with multiple dispatch as its core programming paradigm. Julia supports concurrent, (composable) parallel and distributed computing (with or without using MPI or the built-in corresponding to "OpenMP-style" threads), and direct calling of C and Fortran libraries without glue code. Julia uses a just-in-time (JIT) compiler that is referred to as "just- ahead-of-time" (JAOT) in the Julia community, as Julia compiles all code (by default) to machine code before running it. Julia is garbage-collected, uses eager evaluation, and includes efficient libraries for floating-point calculations, linear algebra, random number generation, and regular expression matching. Many libraries are available, including some (e.g., for fast Fo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ELKI
ELKI (for ''Environment for DeveLoping KDD-Applications Supported by Index-Structures'') is a data mining (KDD, knowledge discovery in databases) software framework developed for use in research and teaching. It was originally at the database systems research unit of Professor Hans-Peter Kriegel at the Ludwig Maximilian University of Munich, Germany, and now continued at the Technical University of Dortmund, Germany. It aims at allowing the development and evaluation of advanced data mining algorithms and their interaction with database index structures. Description The ELKI framework is written in Java and built around a modular architecture. Most currently included algorithms belong to clustering, outlier detection and database indexes. The object-oriented architecture allows the combination of arbitrary algorithms, data types, distance functions, indexes, and evaluation measures. The Java just-in-time compiler optimizes all combinations to a similar extent, making benc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Java (programming Language)
Java is a high-level, class-based, object-oriented programming language that is designed to have as few implementation dependencies as possible. It is a general-purpose programming language intended to let programmers ''write once, run anywhere'' ( WORA), meaning that compiled Java code can run on all platforms that support Java without the need to recompile. Java applications are typically compiled to bytecode that can run on any Java virtual machine (JVM) regardless of the underlying computer architecture. The syntax of Java is similar to C and C++, but has fewer low-level facilities than either of them. The Java runtime provides dynamic capabilities (such as reflection and runtime code modification) that are typically not available in traditional compiled languages. , Java was one of the most popular programming languages in use according to GitHub, particularly for client–server web applications, with a reported 9 million developers. Java was originally developed ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
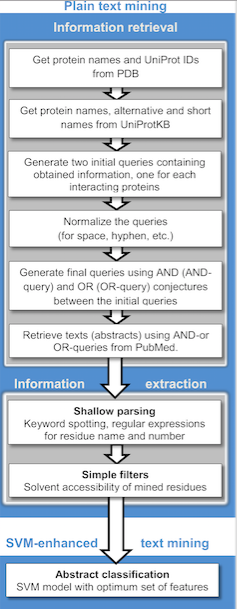
Text Mining
Text mining, also referred to as ''text data mining'', similar to text analytics, is the process of deriving high-quality information from text. It involves "the discovery by computer of new, previously unknown information, by automatically extracting information from different written resources." Written resources may include websites, books, emails, reviews, and articles. High-quality information is typically obtained by devising patterns and trends by means such as statistical pattern learning. According to Hotho et al. (2005) we can distinguish between three different perspectives of text mining: information extraction, data mining, and a KDD (Knowledge Discovery in Databases) process. Text mining usually involves the process of structuring the input text (usually parsing, along with the addition of some derived linguistic features and the removal of others, and subsequent insertion into a database), deriving patterns within the structured data, and finally evaluation and inte ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Interaction Graph
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residues in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Principal Component Analysis
Principal component analysis (PCA) is a popular technique for analyzing large datasets containing a high number of dimensions/features per observation, increasing the interpretability of data while preserving the maximum amount of information, and enabling the visualization of multidimensional data. Formally, PCA is a statistical technique for reducing the dimensionality of a dataset. This is accomplished by linearly transforming the data into a new coordinate system where (most of) the variation in the data can be described with fewer dimensions than the initial data. Many studies use the first two principal components in order to plot the data in two dimensions and to visually identify clusters of closely related data points. Principal component analysis has applications in many fields such as population genetics, microbiome studies, and atmospheric science. The principal components of a collection of points in a real coordinate space are a sequence of p unit vectors, where th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Computational Biology
Computational biology refers to the use of data analysis, mathematical modeling and computational simulations to understand biological systems and relationships. An intersection of computer science, biology, and big data, the field also has foundations in applied mathematics, chemistry, and genetics. It differs from biological computing, a subfield of computer engineering which uses bioengineering to build computers. History Bioinformatics, the analysis of informatics processes in biological systems, began in the early 1970s. At this time, research in artificial intelligence was using network models of the human brain in order to generate new algorithms. This use of biological data pushed biological researchers to use computers to evaluate and compare large data sets in their own field. By 1982, researchers shared information via punch cards. The amount of data grew exponentially by the end of the 1980s, requiring new computational methods for quickly interpreting ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |




.jpg)