Single-cell Transcriptomics on:
[Wikipedia]
[Google]
[Amazon]
Single-cell transcriptomics examines the
 There are several methods available to isolate and amplify cells for single-cell analysis. Low throughput techniques are able to isolate hundreds of cells, are slow, and enable selection. These methods include:
* Micropipetting
There are several methods available to isolate and amplify cells for single-cell analysis. Low throughput techniques are able to isolate hundreds of cells, are slow, and enable selection. These methods include:
* Micropipetting
Cytoplasmic aspiration
*
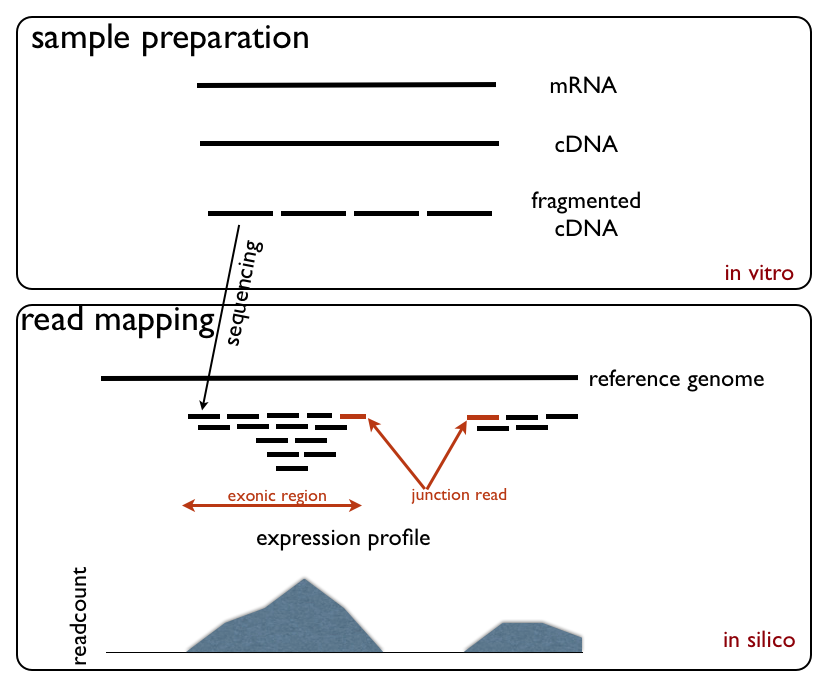 The Single-cell
The Single-cell

 Clustering allows for the formation of subgroups in the cell population. Cells can be clustered by their transcriptomic profile in order to analyse the sub-population structure and identify rare cell types or cell subtypes. Alternatively, genes can be clustered by their expression states in order to identify covarying genes. A combination of both clustering approaches, known as
Clustering allows for the formation of subgroups in the cell population. Cells can be clustered by their transcriptomic profile in order to analyse the sub-population structure and identify rare cell types or cell subtypes. Alternatively, genes can be clustered by their expression states in order to identify covarying genes. A combination of both clustering approaches, known as

 Pseudo-temporal ordering (or trajectory inference) is a technique that aims to infer gene expression dynamics from snapshot single-cell data. The method tries to order the cells in such a way that similar cells are closely positioned to each other. This trajectory of cells can be linear, but can also bifurcate or follow more complex graph structures. The trajectory, therefore, enables the inference of gene expression dynamics and the ordering of cells by their progression through differentiation or response to external stimuli.
The method relies on the assumptions that the cells follow the same path through the process of interest and that their transcriptional state correlates to their progression. The algorithm can be applied to both mixed populations and temporal samples.
More than 50 methods for pseudo-temporal ordering have been developed, and each has its own requirements for prior information (such as starting cells or time course data), detectable topologies, and methodology. An example algorithm is the Monocle algorithm that carries out dimensionality reduction of the data, builds a minimal spanning tree using the transformed data, orders cells in pseudo-time by following the longest connected path of the tree and consequently labels cells by type. Another example is the diffusion pseudotime (DPT) algorithm, which uses a diffusion map and diffusion process. Another class of methods such as MARGARET employ graph partitioning for capturing complex trajectory topologies such as disconnected and multifurcating trajectories.
Pseudo-temporal ordering (or trajectory inference) is a technique that aims to infer gene expression dynamics from snapshot single-cell data. The method tries to order the cells in such a way that similar cells are closely positioned to each other. This trajectory of cells can be linear, but can also bifurcate or follow more complex graph structures. The trajectory, therefore, enables the inference of gene expression dynamics and the ordering of cells by their progression through differentiation or response to external stimuli.
The method relies on the assumptions that the cells follow the same path through the process of interest and that their transcriptional state correlates to their progression. The algorithm can be applied to both mixed populations and temporal samples.
More than 50 methods for pseudo-temporal ordering have been developed, and each has its own requirements for prior information (such as starting cells or time course data), detectable topologies, and methodology. An example algorithm is the Monocle algorithm that carries out dimensionality reduction of the data, builds a minimal spanning tree using the transformed data, orders cells in pseudo-time by following the longest connected path of the tree and consequently labels cells by type. Another example is the diffusion pseudotime (DPT) algorithm, which uses a diffusion map and diffusion process. Another class of methods such as MARGARET employ graph partitioning for capturing complex trajectory topologies such as disconnected and multifurcating trajectories.
Dissecting Tumor Heterogeneity with Single-Cell Transcriptomics
The ultimate single-cell RNA sequencing guide
by single-cell RNA sequencing service provider Single Cell Discoveries. DNA sequencing Molecular biology techniques Biotechnology
gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a ba ...
expression level of individual cells in a given population by simultaneously measuring the RNA concentration (conventionally only messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the p ...
(mRNA)) of hundreds to thousands of genes. Single-cell transcriptomics makes it possible to unravel heterogeneous
Homogeneity and heterogeneity are concepts often used in the sciences and statistics relating to the uniformity of a substance or organism. A material or image that is homogeneous is uniform in composition or character (i.e. color, shape, siz ...
cell populations, reconstruct cellular developmental pathways, and model transcriptional dynamics — all previously masked in bulk RNA sequencing.
Background
The development of high-throughputRNA sequencing
RNA-Seq (named as an abbreviation of RNA sequencing) is a sequencing technique which uses next-generation sequencing (NGS) to reveal the presence and quantity of RNA in a biological sample at a given moment, analyzing the continuously changing ...
(RNA-seq) and microarrays
A microarray is a multiplex lab-on-a-chip. Its purpose is to simultaneously detect the expression of thousands of genes from a sample (e.g. from a tissue). It is a two-dimensional array on a solid substrate—usually a glass slide or silicon ...
has made gene expression analysis a routine. RNA analysis was previously limited to tracing individual transcripts by Northern blot
The northern blot, or RNA blot,Gilbert, S. F. (2000) Developmental Biology, 6th Ed. Sunderland MA, Sinauer Associates. is a technique used in molecular biology research to study gene expression by detection of RNA (or isolated mRNA) in a sample ...
s or quantitative PCR
A real-time polymerase chain reaction (real-time PCR, or qPCR) is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR (i.e., in real ...
. Higher throughput and speed allow researchers to frequently characterize the expression profiles of populations of thousands of cells. The data from bulk assays
An assay is an investigative (analytic) procedure in laboratory medicine, mining, pharmacology, environmental biology and molecular biology for qualitatively assessing or quantitatively measuring the presence, amount, or functional activity of a ...
has led to identifying genes differentially expressed in distinct cell populations, and biomarker
In biomedical contexts, a biomarker, or biological marker, is a measurable indicator of some biological state or condition. Biomarkers are often measured and evaluated using blood, urine, or soft tissues to examine normal biological processes, p ...
discovery.
These studies are limited as they provide measurements for whole tissues and, as a result, show an average expression profile for all the constituent cells. This has a couple of drawbacks. Firstly, different cell types within the same tissue can have distinct roles in multicellular organisms. They often form subpopulations with unique transcriptional profiles. Correlations in the gene expression of the subpopulations can often be missed due to the lack of subpopulation identification. Secondly, bulk assays fail to recognize whether a change in the expression profile is due to a change in regulation or composition — for example if one cell type arises to dominate the population. Lastly, when your goal is to study cellular progression through differentiation, average expression profiles can only order cells by time rather than by developmental stage. Consequently, they cannot show trends in gene expression levels specific to certain stages.
Recent advances in biotechnology allow the measurement of gene expression in hundreds to thousands of individual cells simultaneously. While these breakthroughs in transcriptomics technologies
Transcriptomics technologies are the techniques used to study an organism's transcriptome, the sum of all of its RNA transcripts. The information content of an organism is recorded in the DNA of its genome and expressed through transcription. ...
have enabled the generation of single-cell transcriptomic data, they also presented new computational and analytical challenges. Bioinformaticians can use techniques from bulk RNA-seq for single-cell data. Still, many new computational approaches have had to be designed for this data type to facilitate a complete and detailed study of single-cell expression profiles.
Experimental steps
There is currently no standardized technique to generate single-cell data, all methods must include cell isolation from the population,lysate
Lysis ( ) is the breaking down of the membrane of a cell, often by viral, enzymic, or osmotic (that is, "lytic" ) mechanisms that compromise its integrity. A fluid containing the contents of lysed cells is called a ''lysate''. In molecular bio ...
formation, amplification through reverse transcription and quantification of expression levels. Common techniques for measuring expression are quantitative PCR or RNA-seq.
Isolating single cells
 There are several methods available to isolate and amplify cells for single-cell analysis. Low throughput techniques are able to isolate hundreds of cells, are slow, and enable selection. These methods include:
* Micropipetting
There are several methods available to isolate and amplify cells for single-cell analysis. Low throughput techniques are able to isolate hundreds of cells, are slow, and enable selection. These methods include:
* Micropipetting
Cytoplasmic aspiration
*
Laser capture microdissection
Laser capture microdissection (LCM), also called microdissection, laser microdissection (LMD), or laser-assisted microdissection (LMD or LAM), is a method for isolating specific cells of interest from microscopic regions of tissue/cells/organisms ...
.
High-throughput methods are able to quickly isolate hundreds to tens of thousands of cells. Common techniques include:
* Fluorescence activated cell sorting (FACS)
* Microfluidic
Microfluidics refers to the behavior, precise control, and manipulation of fluids that are geometrically constrained to a small scale (typically sub-millimeter) at which surface forces dominate volumetric forces. It is a multidisciplinary field tha ...
devices
Combining FACS with scRNA-seq has produced optimized protocols such as SORT-seq. A list of studies that utilized SORT-seq can be found here. Moreover, combining microfluidic devices with scRNA-seq has been optimized in 10x Genomics protocols.
Quantitative PCR (qPCR)
To measure the level of expression of each transcript qPCR can be applied. Gene specific primers are used to amplify the corresponding gene as with regular PCR and as a result data is usually only obtained for sample sizes of less than 100 genes. The inclusion ofhousekeeping genes
In molecular biology, housekeeping genes are typically constitutive genes that are required for the maintenance of basic cellular function, and are expressed in all cells of an organism under normal and patho-physiological conditions. Although ...
, whose expression should be
constant under the conditions, is used for normalisation. The most commonly used house keeping genes include GAPDH
Glyceraldehyde 3-phosphate dehydrogenase (abbreviated GAPDH) () is an enzyme of about 37kDa that catalyzes the sixth step of glycolysis and thus serves to break down glucose for energy and carbon molecules. In addition to this long establishe ...
and α-actin
Actin is a family of globular multi-functional proteins that form microfilaments in the cytoskeleton, and the thin filaments in muscle fibrils. It is found in essentially all eukaryotic cells, where it may be present at a concentration of over ...
, although the reliability of normalisation through this process is questionable as there is evidence that the level of expression can vary significantly. Fluorescent dyes
A fluorophore (or fluorochrome, similarly to a chromophore) is a fluorescent chemical compound that can re-emit light upon light excitation. Fluorophores typically contain several combined aromatic groups, or planar or cyclic molecules with sev ...
are used as reporter molecules to detect the PCR product and monitor the progress of the amplification - the increase in fluorescence intensity is proportional to the amplicon
In molecular biology, an amplicon is a piece of DNA or RNA that is the source and/or product of amplification (molecular biology), amplification or DNA replication, replication events. It can be formed artificially, using various methods including ...
concentration. A plot of fluorescence vs. cycle
number is made and a threshold fluorescence level is used to find cycle number at which the plot reaches this value. The cycle number at this point is known as the threshold cycle (Ct) and is measured for each gene.
Single-cell RNA-seq
 The Single-cell
The Single-cell RNA-seq
RNA-Seq (named as an abbreviation of RNA sequencing) is a sequencing technique which uses next-generation sequencing (NGS) to reveal the presence and quantity of RNA in a biological sample at a given moment, analyzing the continuously changing c ...
technique converts a population of RNAs to a library of cDNA fragments. These fragments are sequenced by high-throughput next generation sequencing
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The ...
techniques and the reads are mapped back to the reference genome, providing a count of the number of reads associated with each gene.
Normalisation of RNA-seq data accounts for cell to cell variation in the efficiencies of the cDNA library formation and sequencing. One method relies on the use of extrinsic
In science and engineering, an intrinsic property is a property of a specified subject that exists itself or within the subject. An extrinsic property is not essential or inherent to the subject that is being characterized. For example, mass ...
RNA spike-ins (RNA sequences of known sequence and quantity) that are added in equal quantities to each cell lysate
Lysis ( ) is the breaking down of the membrane of a cell, often by viral, enzymic, or osmotic (that is, "lytic" ) mechanisms that compromise its integrity. A fluid containing the contents of lysed cells is called a ''lysate''. In molecular bio ...
and used to normalise read count by the number of reads mapped to spike-in mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein.
mRNA is ...
.
Another control uses unique molecular identifiers Unique molecular identifiers (UMIs), or molecular barcodes (MBC) are short sequences or molecular "tags" added to DNA fragments in some next generation sequencing library preparation protocols to identify the input DNA molecule. These tags are added ...
(UMIs)-short DNA sequences (6–10nt) that are added to each cDNA before amplification and act as a bar code for each cDNA molecule. Normalisation is achieved by using the count number of unique UMIs associated with each gene to account for differences in amplification efficiency.
A combination of both spike-ins, UMIs and other approaches have been combined for more accurate normalisation.
Considerations
A problem associated with single-cell data occurs in the form of zero inflated gene expression distributions, known as technical dropouts, that are common due to low mRNA concentrations of less-expressed genes that are not captured in the reverse transcription process. The percentage of mRNA molecules in the cell lysate that are detected is often only 10-20%. When using RNA spike-ins for normalisation the assumption is made that the amplification and sequencing efficiencies for theendogenous
Endogenous substances and processes are those that originate from within a living system such as an organism, tissue, or cell.
In contrast, exogenous substances and processes are those that originate from outside of an organism.
For example, es ...
and spike-in RNA are the same. Evidence suggests that this is not the case given fundamental differences in size and features, such as the lack of a polyadenylated tail in spike-ins and therefore shorter length. Additionally, normalisation using UMIs assumes the cDNA library is sequenced to saturation, which is not always the case.
Data analysis
Insights based on single-cell data analysis assume that the input is a matrix of normalised gene expression counts, generated by the approaches outlined above, and can provide opportunities that are not obtainable by bulk. Three main insights provided: #Identification and characterization of cell types and their spatial organisation in time #Inference ofgene regulatory networks
A gene (or genetic) regulatory network (GRN) is a collection of molecular regulators that interact with each other and with other substances in the cell to govern the gene expression levels of mRNA and proteins which, in turn, determine the fun ...
and their strength across individual cells
#Classification of the stochastic
Stochastic (, ) refers to the property of being well described by a random probability distribution. Although stochasticity and randomness are distinct in that the former refers to a modeling approach and the latter refers to phenomena themselv ...
component of transcription
The techniques outlined have been designed to help visualise and explore patterns in the data in order to facilitate the revelation of these three features.
Clustering
 Clustering allows for the formation of subgroups in the cell population. Cells can be clustered by their transcriptomic profile in order to analyse the sub-population structure and identify rare cell types or cell subtypes. Alternatively, genes can be clustered by their expression states in order to identify covarying genes. A combination of both clustering approaches, known as
Clustering allows for the formation of subgroups in the cell population. Cells can be clustered by their transcriptomic profile in order to analyse the sub-population structure and identify rare cell types or cell subtypes. Alternatively, genes can be clustered by their expression states in order to identify covarying genes. A combination of both clustering approaches, known as biclustering Biclustering, block clustering, Co-clustering or two-mode clustering is a data mining technique which allows simultaneous clustering of the rows and columns of a matrix.
The term was first introduced by Boris Mirkin
to name a technique introduce ...
, has been used to simultaneously cluster by genes and cells to find genes that behave similarly within cell clusters.
Clustering methods applied can be K-means clustering, forming disjoint groups or Hierarchical clustering
In data mining and statistics, hierarchical clustering (also called hierarchical cluster analysis or HCA) is a method of cluster analysis that seeks to build a hierarchy of clusters. Strategies for hierarchical clustering generally fall into ...
, forming nested partitions.
Biclustering
Biclustering provides several advantages by improving the resolution of clustering. Genes that are only informative to a subset of cells and are hence only expressed there can be identified through biclustering. Moreover, similarly behaving genes that differentiate one cell cluster from another can be identified using this method.Dimensionality reduction

Dimensionality reduction
Dimensionality reduction, or dimension reduction, is the transformation of data from a high-dimensional space into a low-dimensional space so that the low-dimensional representation retains some meaningful properties of the original data, ideally ...
algorithms such as Principal component analysis
Principal component analysis (PCA) is a popular technique for analyzing large datasets containing a high number of dimensions/features per observation, increasing the interpretability of data while preserving the maximum amount of information, and ...
(PCA) and t-SNE
t-distributed stochastic neighbor embedding (t-SNE) is a statistical method for visualizing high-dimensional data by giving each datapoint a location in a two or three-dimensional map. It is based on Stochastic Neighbor Embedding originally d ...
can be used to simplify data for visualisation and pattern detection by transforming cells from a high to a lower dimensional space
In physics and mathematics, the dimension of a mathematical space (or object) is informally defined as the minimum number of coordinates needed to specify any point within it. Thus, a line has a dimension of one (1D) because only one coordina ...
. The result of this method produces graphs with each cell as a point in a 2-D or 3-D space. Dimensionality reduction is frequently used before clustering as cells in high dimensions can wrongly appear to be close due to distance metrics behaving non-intuitively.
Principal component analysis
The most frequently used technique is PCA, which identifies the directions of largestvariance
In probability theory and statistics, variance is the expectation of the squared deviation of a random variable from its population mean or sample mean. Variance is a measure of dispersion, meaning it is a measure of how far a set of numbers ...
principal components and transforms the data so that the first principal component has the largest possible variance, and successive principle components in turn each have the highest variance possible while remaining orthogonal to the preceding components. The contribution each gene makes to each component is used to infer which genes are contributing the most to variance in the population and are involved in differentiating different subpopulations.
Differential expression
Detecting differences in gene expression level between two populations is used both single-cell and bulk transcriptomic data. Specialised methods have been designed for single-cell data that considers single cell features such as technical dropouts and shape of the distribution e.g.Bimodal
In statistics, a multimodal distribution is a probability distribution with more than one mode (statistics), mode. These appear as distinct peaks (local maxima) in the probability density function, as shown in Figures 1 and 2. Categorical, ...
vs. unimodal
In mathematics, unimodality means possessing a unique mode. More generally, unimodality means there is only a single highest value, somehow defined, of some mathematical object.
Unimodal probability distribution
In statistics, a unimodal pr ...
.
Gene ontology enrichment
Gene ontology
The Gene Ontology (GO) is a major bioinformatics initiative to unify the representation of gene and gene product attributes across all species. More specifically, the project aims to: 1) maintain and develop its controlled vocabulary of gene and g ...
terms describe gene functions and the relationships between those functions into three classes:
#Molecular function
#Cellular component
#Biological process
Gene Ontology (GO) term enrichment is a technique used to identify which GO terms are over-represented or under-represented in a given set of genes. In single-cell analysis input list of genes of interest can be selected based on differentially expressed genes or groups of genes generated from biclustering. The number of genes annotated to a GO term in the input list is normalised against the number of genes annotated to a GO term in the background set of all genes in genome to determine statistical significance.
Pseudotemporal ordering
Network inference
Gene regulatory network inference is a technique that aims to construct a network, shown as a graph, in which thenodes
In general, a node is a localized swelling (a "knot") or a point of intersection (a Vertex (graph theory), vertex).
Node may refer to:
In mathematics
*Vertex (graph theory), a vertex in a mathematical graph
*Vertex (geometry), a point where two ...
represent the genes and edges indicate co-regulatory interactions. The method relies on the assumption that a strong statistical relationship between the expression of genes is an indication of a potential functional relationship. The most commonly used method to measure the strength of a statistical relationship is correlation
In statistics, correlation or dependence is any statistical relationship, whether causal or not, between two random variables or bivariate data. Although in the broadest sense, "correlation" may indicate any type of association, in statistics ...
. However, correlation fails to identify non-linear
In mathematics and science, a nonlinear system is a system in which the change of the output is not proportional to the change of the input. Nonlinear problems are of interest to engineers, biologists, physicists, mathematicians, and many other ...
relationships and mutual information
In probability theory and information theory, the mutual information (MI) of two random variables is a measure of the mutual dependence between the two variables. More specifically, it quantifies the " amount of information" (in units such ...
is used as an alternative. Gene clusters linked in a network signify genes that undergo coordinated changes in expression.
Integration
The presence or strength of technical effects and the types of cells observed often differ in single-cell transcriptomics datasets generated using different experimental protocols and under different conditions. This difference results in strong batch effects that may bias the findings of statistical methods applied across batches, particularly in the presence ofconfounding
In statistics, a confounder (also confounding variable, confounding factor, extraneous determinant or lurking variable) is a variable that influences both the dependent variable and independent variable, causing a spurious association. Con ...
.
As a result of the aforementioned properties of single-cell transcriptomic data, batch correction methods developed for bulk sequencing data were observed to perform poorly. Consequently, researchers developed statistical methods to correct for batch effects that are robust to the properties of single-cell transcriptomic data to integrate data from different sources or experimental batches. Laleh Haghverdi performed foundational work in formulating the use of mutual nearest neighbors between each batch to define batch correction vectors. With these vectors, you can merge datasets that each include at least one shared cell type. An orthogonal approach involves the projection of each dataset onto a shared low-dimensional space using canonical correlation analysis
In statistics, canonical-correlation analysis (CCA), also called canonical variates analysis, is a way of inferring information from cross-covariance matrices. If we have two vectors ''X'' = (''X''1, ..., ''X'n'') and ''Y' ...
. Mutual nearest neighbors and canonical correlation analysis have also been combined to define integration "anchors" comprising reference cells in one dataset, to which query cells in another dataset are normalized.
See also
*RNA-Seq
RNA-Seq (named as an abbreviation of RNA sequencing) is a sequencing technique which uses next-generation sequencing (NGS) to reveal the presence and quantity of RNA in a biological sample at a given moment, analyzing the continuously changing c ...
* Single-cell analysis
In the field of cellular biology, single-cell analysis is the study of genomics, transcriptomics, proteomics, metabolomics and cell–cell interactions at the single cell level. The concept of single-cell analysis originated in th ...
* Single-cell sequencing
* Transcriptome
The transcriptome is the set of all RNA transcripts, including coding and non-coding, in an individual or a population of cells. The term can also sometimes be used to refer to all RNAs, or just mRNA, depending on the particular experiment. The t ...
* Transcriptomics
Transcriptomics technologies are the techniques used to study an organism's transcriptome, the sum of all of its RNA transcripts. The information content of an organism is recorded in the DNA of its genome and expressed through transcription. He ...
References
{{Reflist, 35emExternal links
Dissecting Tumor Heterogeneity with Single-Cell Transcriptomics
The ultimate single-cell RNA sequencing guide
by single-cell RNA sequencing service provider Single Cell Discoveries. DNA sequencing Molecular biology techniques Biotechnology