|
Single-cell Transcriptomics
Single-cell transcriptomics examines the gene expression level of individual cells in a given population by simultaneously measuring the RNA concentration (conventionally only messenger RNA (mRNA)) of hundreds to thousands of genes. Single-cell transcriptomics makes it possible to unravel heterogeneous cell populations, reconstruct cellular developmental pathways, and model transcriptional dynamics — all previously masked in bulk RNA sequencing. Background The development of high-throughput RNA sequencing (RNA-seq) and microarrays has made gene expression analysis a routine. RNA analysis was previously limited to tracing individual transcripts by Northern blots or quantitative PCR. Higher throughput and speed allow researchers to frequently characterize the expression profiles of populations of thousands of cells. The data from bulk assays has led to identifying genes differentially expressed in distinct cell populations, and biomarker discovery. These studies are limite ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gene
In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and noncoding genes. During gene expression, the DNA is first copied into RNA. The RNA can be directly functional or be the intermediate template for a protein that performs a function. The transmission of genes to an organism's offspring is the basis of the inheritance of phenotypic traits. These genes make up different DNA sequences called genotypes. Genotypes along with environmental and developmental factors determine what the phenotypes will be. Most biological traits are under the influence of polygenes (many different genes) as well as g ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Laser Capture Microdissection
Laser capture microdissection (LCM), also called microdissection, laser microdissection (LMD), or laser-assisted microdissection (LMD or LAM), is a method for isolating specific cells of interest from microscopic regions of tissue/cells/organisms (dissection on a microscopic scale with the help of a laser). Principle Laser-capture microdissection (LCM) is a method to procure subpopulations of tissue cells under direct microscopic visualization. LCM technology can harvest the cells of interest directly or can isolate specific cells by cutting away unwanted cells to give histologically pure enriched cell populations. A variety of downstream applications exist: DNA genotyping and loss of heterozygosity (LOH) analysis, RNA transcript profiling, cDNA library generation, proteomics discovery and signal-pathway profiling. The total time required to carry out this protocol is typically 1–1.5 h. Extraction A laser is coupled into a microscope and focuses onto the tissue on the slide. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA-Seq
RNA-Seq (named as an abbreviation of RNA sequencing) is a sequencing technique which uses next-generation sequencing (NGS) to reveal the presence and quantity of RNA in a biological sample at a given moment, analyzing the continuously changing cellular transcriptome. Specifically, RNA-Seq facilitates the ability to look at alternative gene spliced transcripts, post-transcriptional modifications, gene fusion, mutations/ SNPs and changes in gene expression over time, or differences in gene expression in different groups or treatments. In addition to mRNA transcripts, RNA-Seq can look at different populations of RNA to include total RNA, small RNA, such as miRNA, tRNA, and ribosomal profiling. RNA-Seq can also be used to determine exon/intron boundaries and verify or amend previously annotated 5' and 3' gene boundaries. Recent advances in RNA-Seq include single cell sequencing, in situ sequencing of fixed tissue, and native RNA molecule sequencing with single-molecule real- ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
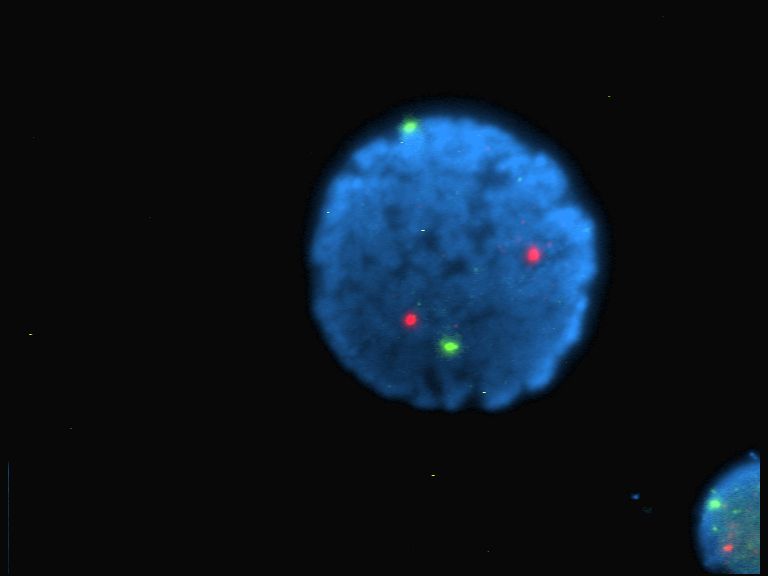
Single Cell Sequencing
Single-cell sequencing examines the sequence information from individual cells with optimized next-generation sequencing technologies, providing a higher resolution of cellular differences and a better understanding of the function of an individual cell in the context of its microenvironment. For example, in cancer, sequencing the DNA of individual cells can give information about mutations carried by small populations of cells. In development, sequencing the RNAs expressed by individual cells can give insight into the existence and behavior of different cell types. In microbial systems, a population of the same species can appear genetically clonal. Still, single-cell sequencing of RNA or epigenetic modifications can reveal cell-to-cell variability that may help populations rapidly adapt to survive in changing environments. Background A typical human cell consists of about 2 x 3.3 billion base pairs of DNA and 600 million mRNA bases. Usually, a mix of millions of cells is used ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNA Seq Experiment
Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and deoxyribonucleic acid ( DNA) are nucleic acids. Along with lipids, proteins, and carbohydrates, nucleic acids constitute one of the four major macromolecules essential for all known forms of life. Like DNA, RNA is assembled as a chain of nucleotides, but unlike DNA, RNA is found in nature as a single strand folded onto itself, rather than a paired double strand. Cellular organisms use messenger RNA (''mRNA'') to convey genetic information (using the nitrogenous bases of guanine, uracil, adenine, and cytosine, denoted by the letters G, U, A, and C) that directs synthesis of specific proteins. Many viruses encode their genetic information using an RNA genome. Some RNA molecules play an active role within cells by catalyzing biological reactions, controlling gene expression, or sensing and communicating responses to cellular signa ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Amplicon
In molecular biology, an amplicon is a piece of DNA or RNA that is the source and/or product of amplification or replication events. It can be formed artificially, using various methods including polymerase chain reactions (PCR) or ligase chain reactions (LCR), or naturally through gene duplication. In this context, ''amplification'' refers to the production of one or more copies of a genetic fragment or target sequence, specifically the amplicon. As it refers to the product of an amplification reaction, ''amplicon'' is used interchangeably with common laboratory terms, such as "PCR product." Artificial amplification is used in research, forensics, and medicine for purposes that include detection and quantification of infectious agents, identification of human remains, and extracting genotypes from human hair. Natural gene duplication plays a major role in evolution. It is also implicated in several forms of human cancer including primary mediastinal B cell lymphoma and Hodgkin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reporter Gene
In molecular biology, a reporter gene (often simply reporter) is a gene that researchers attach to a regulatory sequence of another gene of interest in bacteria, cell culture, animals or plants. Such genes are called reporters because the characteristics they confer on organisms expressing them are easily identified and measured, or because they are selectable markers. Reporter genes are often used as an indication of whether a certain gene has been taken up by or expressed in the cell or organism population. Common reporter genes To introduce a reporter gene into an organism, scientists place the reporter gene and the gene of interest in the same DNA construct to be inserted into the cell or organism. For bacteria or prokaryotic cells in culture, this is usually in the form of a circular DNA molecule called a plasmid. For viruses, this is known as a viral vector. It is important to use a reporter gene that is not natively expressed in the cell or organism under study, since ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fluorescent Dyes
A fluorophore (or fluorochrome, similarly to a chromophore) is a fluorescent chemical compound that can re-emit light upon light excitation. Fluorophores typically contain several combined aromatic groups, or planar or cyclic molecules with several π bonds. Fluorophores are sometimes used alone, as a tracer in fluids, as a dye for staining of certain structures, as a substrate of enzymes, or as a probe or indicator (when its fluorescence is affected by environmental aspects such as polarity or ions). More generally they are covalently bonded to a macromolecule, serving as a marker (or dye, or tag, or reporter) for affine or bioactive reagents (antibodies, peptides, nucleic acids). Fluorophores are notably used to stain tissues, cells, or materials in a variety of analytical methods, i.e., fluorescent imaging and spectroscopy. Fluorescein, via its amine-reactive isothiocyanate derivative fluorescein isothiocyanate (FITC), has been one of the most popular fluorophores. From an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Actin
Actin is a protein family, family of Globular protein, globular multi-functional proteins that form microfilaments in the cytoskeleton, and the thin filaments in myofibril, muscle fibrils. It is found in essentially all Eukaryote, eukaryotic cells, where it may be present at a concentration of over 100 micromolar, μM; its mass is roughly 42 kDa, with a diameter of 4 to 7 nm. An actin protein is the monomeric Protein subunit, subunit of two types of filaments in cells: microfilaments, one of the three major components of the cytoskeleton, and thin filaments, part of the Muscle contraction, contractile apparatus in muscle cells. It can be present as either a free monomer called G-actin (globular) or as part of a linear polymer microfilament called F-actin (filamentous), both of which are essential for such important cellular functions as the Motility, mobility and contraction of cell (biology), cells during cell division. Actin participates in many important cellular pr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
GAPDH
Glyceraldehyde 3-phosphate dehydrogenase (abbreviated GAPDH) () is an enzyme of about 37kDa that catalyzes the sixth step of glycolysis and thus serves to break down glucose for energy and carbon molecules. In addition to this long established metabolic function, GAPDH has recently been implicated in several non-metabolic processes, including transcription activation, initiation of apoptosis, ER to Golgi vesicle shuttling, and fast axonal, or axoplasmic transport. In sperm, a testis-specific isoenzyme GAPDHS is expressed. Structure Under normal cellular conditions, cytoplasmic GAPDH exists primarily as a tetramer. This form is composed of four identical 37-kDa subunits containing a single catalytic thiol group each and critical to the enzyme's catalytic function. Nuclear GAPDH has increased isoelectric point (pI) of pH 8.3–8.7. Of note, the cysteine residue C152 in the enzyme's active site is required for the induction of apoptosis by oxidative stress. Notably, pos ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Housekeeping Genes
In molecular biology, housekeeping genes are typically constitutive genes that are required for the maintenance of basic cellular function, and are expressed in all cells of an organism under normal and patho-physiological conditions. Although some housekeeping genes are expressed at relatively constant rates in most non-pathological situations, the expression of other housekeeping genes may vary depending on experimental conditions. The origin of the term "housekeeping gene" remains obscure. Literature from 1976 used the term to describe specifically tRNA and rRNA. For experimental purposes, the expression of one or multiple housekeeping genes is used as a reference point for the analysis of expression levels of other genes. The key criterion for the use of a housekeeping gene in this manner is that the chosen housekeeping gene is uniformly expressed with low variance under both control and experimental conditions. Validation of housekeeping genes should be performed before the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is a method widely used to rapidly make millions to billions of copies (complete or partial) of a specific DNA sample, allowing scientists to take a very small sample of DNA and amplify it (or a part of it) to a large enough amount to study in detail. PCR was invented in 1983 by the American biochemist Kary Mullis at Cetus Corporation; Mullis and biochemist Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993. PCR is fundamental to many of the procedures used in genetic testing and research, including analysis of ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory research for a broad variety of applications including biomedical research ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
_B2.jpg)