primary transcript on:
[Wikipedia]
[Google]
[Amazon]
 A primary transcript is the single-stranded ribonucleic acid (
A primary transcript is the single-stranded ribonucleic acid (
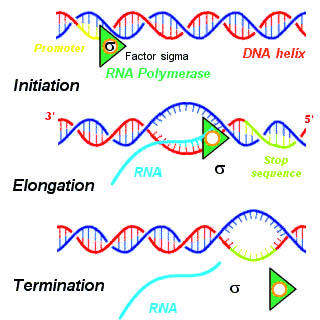 In eukaryotes, three kinds of RNA— rRNA, tRNA, and mRNA—are produced based on the activity of three distinct RNA polymerases, whereas, in
In eukaryotes, three kinds of RNA— rRNA, tRNA, and mRNA—are produced based on the activity of three distinct RNA polymerases, whereas, in
 Inhibition of RNA polymerase activity can also be regulated by DNA sequences called silencers. Like enhancers, silencers may be located at locations farther up or downstream from the genes they regulate. These DNA sequences bind to factors that contribute to the destabilization of the initiation complex required to activate RNA polymerase, and therefore inhibit transcription.
Histone modification by transcription factors is another key regulatory factor for transcription by RNA polymerase. In general, factors that lead to histone acetylation activate transcription while factors that lead to histone deacetylation inhibit transcription. Acetylation of histones induces repulsion between negative components within nucleosomes, allowing for RNA polymerase access. Deacetylation of histones stabilizes tightly coiled nucleosomes, inhibiting RNA polymerase access. In addition to acetylation patterns of histones, methylation patterns at promoter regions of DNA can regulate RNA polymerase access to a given template. RNA polymerase is often incapable of synthesizing a primary transcript if the targeted gene's promoter region contains specific methylated cytosines— residues that hinder binding of transcription-activating factors and recruit other enzymes to stabilize a tightly bound nucleosome structure, excluding access to RNA polymerase and preventing the production of primary transcripts.
Inhibition of RNA polymerase activity can also be regulated by DNA sequences called silencers. Like enhancers, silencers may be located at locations farther up or downstream from the genes they regulate. These DNA sequences bind to factors that contribute to the destabilization of the initiation complex required to activate RNA polymerase, and therefore inhibit transcription.
Histone modification by transcription factors is another key regulatory factor for transcription by RNA polymerase. In general, factors that lead to histone acetylation activate transcription while factors that lead to histone deacetylation inhibit transcription. Acetylation of histones induces repulsion between negative components within nucleosomes, allowing for RNA polymerase access. Deacetylation of histones stabilizes tightly coiled nucleosomes, inhibiting RNA polymerase access. In addition to acetylation patterns of histones, methylation patterns at promoter regions of DNA can regulate RNA polymerase access to a given template. RNA polymerase is often incapable of synthesizing a primary transcript if the targeted gene's promoter region contains specific methylated cytosines— residues that hinder binding of transcription-activating factors and recruit other enzymes to stabilize a tightly bound nucleosome structure, excluding access to RNA polymerase and preventing the production of primary transcripts.
 The effect of alternative splicing in gene expression can be seen in complex eukaryotes which have a fixed number of genes in their genome yet produce much larger numbers of different gene products. Most eukaryotic pre-mRNA transcripts contain multiple introns and exons. The various possible combinations of 5' and 3' splice sites in a pre-mRNA can lead to different excision and combination of exons while the introns are eliminated from the mature mRNA. Thus, various kinds of mature mRNAs are generated. Alternative splicing takes place in a large protein complex called the spliceosome. Alternative splicing is crucial for tissue-specific and developmental regulation in gene expression. Alternative splicing can be affected by various factors, including mutations such as chromosomal translocation.
In prokaryotes, splicing is done by autocatalytic cleavage or by endolytic cleavage. Autocatalytic cleavages, in which no proteins are involved, are usually reserved for sections that code for rRNA, whereas endolytic cleavage corresponds to tRNA precursors.
The effect of alternative splicing in gene expression can be seen in complex eukaryotes which have a fixed number of genes in their genome yet produce much larger numbers of different gene products. Most eukaryotic pre-mRNA transcripts contain multiple introns and exons. The various possible combinations of 5' and 3' splice sites in a pre-mRNA can lead to different excision and combination of exons while the introns are eliminated from the mature mRNA. Thus, various kinds of mature mRNAs are generated. Alternative splicing takes place in a large protein complex called the spliceosome. Alternative splicing is crucial for tissue-specific and developmental regulation in gene expression. Alternative splicing can be affected by various factors, including mutations such as chromosomal translocation.
In prokaryotes, splicing is done by autocatalytic cleavage or by endolytic cleavage. Autocatalytic cleavages, in which no proteins are involved, are usually reserved for sections that code for rRNA, whereas endolytic cleavage corresponds to tRNA precursors.
Scienceden.com RNA Article
{{Nucleic acids RNA
RNA
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself (non-coding RNA) or by forming a template for the production of proteins (messenger RNA). RNA and deoxyrib ...
) product synthesized by transcription of DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
, and processed to yield various mature RNA products such as mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of Protein biosynthesis, synthesizing a protein.
mRNA is ...
s, tRNAs, and rRNAs. The primary transcripts designated to be mRNAs are modified in preparation for translation
Translation is the communication of the semantics, meaning of a #Source and target languages, source-language text by means of an Dynamic and formal equivalence, equivalent #Source and target languages, target-language text. The English la ...
. For example, a precursor mRNA (pre-mRNA) is a type of primary transcript that becomes a messenger RNA (mRNA) after processing.
Pre-mRNA is synthesized from a DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
template in the cell nucleus
The cell nucleus (; : nuclei) is a membrane-bound organelle found in eukaryote, eukaryotic cell (biology), cells. Eukaryotic cells usually have a single nucleus, but a few cell types, such as mammalian red blood cells, have #Anucleated_cells, ...
by transcription. Pre-mRNA comprises the bulk of heterogeneous nuclear RNA (hnRNA). Once pre-mRNA has been completely processed, it is termed " mature messenger RNA", or simply "messenger RNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
". The term hnRNA is often used as a synonym for pre-mRNA, although, in the strict sense, hnRNA may include nuclear RNA transcripts that do not end up as cytoplasmic mRNA.
There are several steps contributing to the production of primary transcripts. All these steps involve a series of interactions to initiate and complete the transcription of DNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
in the nucleus of eukaryotes
The eukaryotes ( ) constitute the domain of Eukaryota or Eukarya, organisms whose cells have a membrane-bound nucleus. All animals, plants, fungi, seaweeds, and many unicellular organisms are eukaryotes. They constitute a major group of ...
. Certain factors play key roles in the activation and inhibition of transcription, where they regulate primary transcript production. Transcription produces primary transcripts that are further modified by several processes. These processes include the 5' cap, 3'-polyadenylation, and alternative splicing
Alternative splicing, alternative RNA splicing, or differential splicing, is an alternative RNA splicing, splicing process during gene expression that allows a single gene to produce different splice variants. For example, some exons of a gene ma ...
. In particular, alternative splicing directly contributes to the diversity of mRNA found in cells. The modifications of primary transcripts have been further studied in research seeking greater knowledge of the role and significance of these transcripts. Experimental studies based on molecular changes to primary transcripts and the processes before and after transcription have led to greater understanding of diseases involving primary transcripts.
Production
The steps contributing to the production of primary transcripts involve a series of molecular interactions that initiate transcription of DNA within a cell's nucleus. Based on the needs of a given cell, certain DNA sequences are transcribed to produce a variety of RNA products to be translated into functional proteins for cellular use. To initiate the transcription process in a cell's nucleus, DNA double helices are unwound andhydrogen bond
In chemistry, a hydrogen bond (H-bond) is a specific type of molecular interaction that exhibits partial covalent character and cannot be described as a purely electrostatic force. It occurs when a hydrogen (H) atom, Covalent bond, covalently b ...
s connecting compatible nucleic acids of DNA are broken to produce two unconnected single DNA strands. One strand of the DNA template is used for transcription of the single-stranded primary transcript mRNA. This DNA strand is bound by an RNA polymerase
In molecular biology, RNA polymerase (abbreviated RNAP or RNApol), or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that catalyzes the chemical reactions that synthesize RNA from a DNA template.
Using the e ...
at the promoter region of the DNA.
 In eukaryotes, three kinds of RNA— rRNA, tRNA, and mRNA—are produced based on the activity of three distinct RNA polymerases, whereas, in
In eukaryotes, three kinds of RNA— rRNA, tRNA, and mRNA—are produced based on the activity of three distinct RNA polymerases, whereas, in prokaryotes
A prokaryote (; less commonly spelled procaryote) is a single-celled organism whose cell lacks a nucleus and other membrane-bound organelles. The word ''prokaryote'' comes from the Ancient Greek (), meaning 'before', and (), meaning 'nut' ...
, only one RNA polymerase exists to create all kinds of RNA molecules. RNA polymerase II of eukaryotes transcribes the primary transcript, a transcript destined to be processed into mRNA, from the antisense DNA template in the 5' to 3' direction, and this newly synthesized primary transcript is complementary to the antisense strand of DNA. RNA polymerase II constructs the primary transcript using a set of four specific ribonucleoside monophosphate residues ( adenosine monophosphate (AMP), cytidine monophosphate (CMP), guanosine monophosphate (GMP), and uridine monophosphate (UMP)) that are added continuously to the 3' hydroxyl group on the 3' end of the growing mRNA.
Studies of primary transcripts produced by RNA polymerase II reveal that an average primary transcript is 7,000 nucleotide
Nucleotides are Organic compound, organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both o ...
s in length, with some growing as long as 20,000 nucleotides in length. The inclusion of both exon
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequence ...
and intron
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e., a region inside a gene."The notion of the cistron .e., gen ...
sequences within primary transcripts explains the size difference between larger primary transcripts and smaller, mature mRNA ready for translation into protein.
Regulation
A number of factors contribute to the activation and inhibition of transcription and therefore regulate the production of primary transcripts from a given DNA template. Activation of RNA polymerase activity to produce primary transcripts is often controlled by sequences of DNA called enhancers. Transcription factors, proteins that bind to DNA elements to either activate or repress transcription, bind to enhancers and recruit enzymes that alternucleosome
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone, histone proteins and resembles thread wrapped around a bobbin, spool. The nucleosome ...
components, causing DNA to be either more or less accessible to RNA polymerase. The unique combinations of either activating or inhibiting transcription factors that bind to enhancer DNA regions determine whether or not the gene that enhancer interacts with is activated for transcription or not. Activation of transcription depends on whether or not the transcription elongation complex, itself consisting of a variety of transcription factors, can induce RNA polymerase to dissociate from the Mediator complex that connects an enhancer region to the promoter. R-loops
R-loops are formed during transcription. An R-loop is a three-stranded nucleic acid structure containing a DNA-RNA hybrid region and an associated non-template single-stranded DNA. Actively transcribed regions ofDNA
Deoxyribonucleic acid (; DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of al ...
often form R-loops that are vulnerable to DNA damage. Introns reduce R-loop formation and DNA damage in highly expressed yeast genes.
Transcription stress
DNA damages arise in each cell, every day, with the number of damages in each cell reaching tens to hundreds of thousands, and such DNA damages can impede primary transcription. The process ofgene expression
Gene expression is the process (including its Regulation of gene expression, regulation) by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, proteins or non-coding RNA, ...
itself is a source of endogenous DNA damages resulting from the susceptibility of single-stranded DNA to damage. Other sources of DNA damage are conflicts of the primary transcription machinery with the DNA replication
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all life, living organisms, acting as the most essential part of heredity, biolog ...
machinery, and the activity of certain enzymes such as topoisomerases and base excision repair enzymes. Even though these processes are tightly regulated and are usually accurate, occasionally they can make mistakes and leave behind DNA breaks that drive chromosomal rearrangements or cell death.
RNA processing
Transcription, a highly regulated phase in gene expression, produces primary transcripts. However, transcription is only the first step which should be followed by many modifications that yield functional forms of RNAs. Otherwise stated, the newly synthesized primary transcripts are modified in several ways to be converted to their mature, functional forms to produce different proteins and RNAs such as mRNA, tRNA, and rRNA.Processing
The basic primary transcript modification process is similar for tRNA and rRNA in both eukaryotic and prokaryotic cells. On the other hand, primary transcript processing varies in mRNAs of prokaryotic and eukaryotic cells. For example, some prokaryotic bacterial mRNAs serve as templates for synthesis of proteins at the same time they are being produced via transcription. Alternatively, pre-mRNA of eukaryotic cells undergo a wide range of modifications prior to their transport from the nucleus to cytoplasm where their mature forms are translated. These modifications are responsible for the different types of encoded messages that lead to translation of various types of products. Furthermore, primary transcript processing provides a control for gene expression as well as a regulatory mechanism for the degradation rates of mRNAs. The processing of pre-mRNA in eukaryotic cells includes 5' capping, 3' polyadenylation, andalternative splicing
Alternative splicing, alternative RNA splicing, or differential splicing, is an alternative RNA splicing, splicing process during gene expression that allows a single gene to produce different splice variants. For example, some exons of a gene ma ...
.
5' capping
Shortly after transcription is initiated in eukaryotes, a pre-mRNA's 5' end is modified by the addition of a 7-methylguanosine cap, also known as a 5' cap. The 5' capping modification is initiated by the addition of a GTP to the 5' terminal nucleotide of the pre-mRNA in reverse orientation followed by the addition of methyl groups to the G residue. 5' capping is essential for the production of functional mRNAs since the 5' cap is responsible for aligning the mRNA with the ribosome during translation.Polyadenylation
In eukaryotes, polyadenylation further modifies pre-mRNAs during which a structure called the poly-A tail is added. Signals for polyadenylation, which include several RNA sequence elements, are detected by a group of proteins which signal the addition of the poly-A tail (approximately 200 nucleotides in length). The polyadenylation reaction provides a signal for the end of transcription and this reaction ends approximately a few hundred nucleotides downstream from the poly-A tail location.Alternative splicing
Eukaryotic pre-mRNAs have their introns spliced out by spliceosomes made up of small nuclear ribonucleoproteins. In complex eukaryotic cells, one primary transcript is able to prepare large amounts of mature mRNAs due to alternative splicing. Alternative splicing is regulated so that each mature mRNA may encode a multiplicity of proteins. The effect of alternative splicing in gene expression can be seen in complex eukaryotes which have a fixed number of genes in their genome yet produce much larger numbers of different gene products. Most eukaryotic pre-mRNA transcripts contain multiple introns and exons. The various possible combinations of 5' and 3' splice sites in a pre-mRNA can lead to different excision and combination of exons while the introns are eliminated from the mature mRNA. Thus, various kinds of mature mRNAs are generated. Alternative splicing takes place in a large protein complex called the spliceosome. Alternative splicing is crucial for tissue-specific and developmental regulation in gene expression. Alternative splicing can be affected by various factors, including mutations such as chromosomal translocation.
In prokaryotes, splicing is done by autocatalytic cleavage or by endolytic cleavage. Autocatalytic cleavages, in which no proteins are involved, are usually reserved for sections that code for rRNA, whereas endolytic cleavage corresponds to tRNA precursors.
The effect of alternative splicing in gene expression can be seen in complex eukaryotes which have a fixed number of genes in their genome yet produce much larger numbers of different gene products. Most eukaryotic pre-mRNA transcripts contain multiple introns and exons. The various possible combinations of 5' and 3' splice sites in a pre-mRNA can lead to different excision and combination of exons while the introns are eliminated from the mature mRNA. Thus, various kinds of mature mRNAs are generated. Alternative splicing takes place in a large protein complex called the spliceosome. Alternative splicing is crucial for tissue-specific and developmental regulation in gene expression. Alternative splicing can be affected by various factors, including mutations such as chromosomal translocation.
In prokaryotes, splicing is done by autocatalytic cleavage or by endolytic cleavage. Autocatalytic cleavages, in which no proteins are involved, are usually reserved for sections that code for rRNA, whereas endolytic cleavage corresponds to tRNA precursors.
Research
5- Fluorouracil (FUra) exposure in methotrexate-resistant KB cells led to a two-fold reduction in total dihydrofolate reductase (DHFR) mRNA levels, while the level of DHFR pre-mRNA with certain introns remained unaffected. The half-life of DHFR mRNA or pre-mRNA did not change significantly, but the transition rate of DHFR RNA from the nucleus to the cytoplasm decreased, suggesting that FUra may influence mRNA processing and/or nuclear DHFR mRNA stability. In '' Drosophila'' and ''Aedes
''Aedes'' (also known as the tiger mosquito) is a genus of mosquitoes originally found in tropical and subtropical zones, but now found on all continents except Antarctica. Some species have been spread by human activity: ''Aedes albopictus'', ...
'', hnRNA (pre-mRNA) size was larger in ''Aedes'' due to its larger genome, despite both species producing mature mRNA of similar size and sequence complexity. This indicates that hnRNA size increases with genome size.
In HeLa cells, spliceosome groups on pre-mRNA were found to form within nuclear speckles, with this formation being temperature-dependent and influenced by specific RNA sequences. Pre-mRNA targeting and splicing factor loading in speckles were critical for spliceosome group formation, resulting in a speckled pattern.
Recruiting pre-mRNA to nuclear speckles significantly increased splicing efficiency and protein levels, indicating that proximity to speckles enhances splicing efficiency.
Related diseases
Research has also led to greater knowledge about certain diseases related to changes within primary transcripts. One study involvedestrogen receptor
Estrogen receptors (ERs) are proteins found in cell (biology), cells that function as receptor (biochemistry), receptors for the hormone estrogen (17β-estradiol). There are two main classes of ERs. The first includes the intracellular estrogen ...
s and differential splicing. The article entitled, "Alternative splicing of the human estrogen receptor alpha primary transcript: mechanisms of exon skipping" by Paola Ferro, Alessandra Forlani, Marco Muselli and Ulrich Pfeffer from the laboratory of Molecular Oncology at National Cancer Research Institute in Genoa, Italy, explains that 1785 nucleotides of the region in the DNA that codes for the estrogen receptor alpha (ER-alpha) are spread over a region that holds more than 300,000 nucleotides in the primary transcript. Splicing of this pre-mRNA frequently leads to variants or different kinds of the mRNA lacking one or more exons or regions necessary for coding proteins. These variants have been associated with breast cancer
Breast cancer is a cancer that develops from breast tissue. Signs of breast cancer may include a Breast lump, lump in the breast, a change in breast shape, dimpling of the skin, Milk-rejection sign, milk rejection, fluid coming from the nipp ...
progression. In the life cycle of retroviruses, proviral DNA is incorporated in transcription of the DNA of the cell being infected. Since retroviruses need to change their pre-mRNA into DNA so that this DNA can be integrated within the DNA of the host it is affecting, the formation of that DNA template is a vital step for retrovirus replication. Cell type, the differentiation or changed state of the cell, and the physiological state of the cell, result in a significant change in the availability and activity of certain factors necessary for transcription. These variables create a wide range of viral gene expression. For example, tissue culture cells actively producing infectious virions of avian or murine leukemia viruses (ASLV or MLV) contain such high levels of viral RNA that 5–10% of the mRNA in a cell can be of viral origin. This shows that the primary transcripts produced by these retroviruses do not always follow the normal path to protein production and convert back into DNA in order to multiply and expand.
See also
* ''cis''-splicing * Outron * ''trans''-splicing *Transcription (biology)
Transcription is the process of copying a segment of DNA into RNA for the purpose of gene expression. Some segments of DNA are transcribed into RNA molecules that can encode proteins, called messenger RNA (mRNA). Other segments of DNA are transc ...
* Transcriptome
References
External links
Scienceden.com RNA Article
{{Nucleic acids RNA