|
Loss-of-function
In biology, a mutation is an alteration in the nucleic acid sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mitosis, or meiosis or other types of damage to DNA (such as pyrimidine dimers caused by exposure to ultraviolet radiation), which then may undergo error-prone repair (especially microhomology-mediated end joining), cause an error during other forms of repair, or cause an error during replication ( translesion synthesis). Mutations may also result from substitution, insertion or deletion of segments of DNA due to mobile genetic elements. Mutations may or may not produce detectable changes in the observable characteristics (phenotype) of an organism. Mutations play a part in both normal and abnormal biological processes including: evolution, cancer, and the development of the immune system, including junctional diversity. Mutation is the ult ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Meiosis
Meiosis () is a special type of cell division of germ cells in sexually-reproducing organisms that produces the gametes, the sperm or egg cells. It involves two rounds of division that ultimately result in four cells, each with only one copy of each chromosome (haploid). Additionally, prior to the division, genetic material from the paternal and maternal copies of each chromosome is crossed over, creating new combinations of code on each chromosome. Later on, during fertilisation, the haploid cells produced by meiosis from a male and a female will fuse to create a zygote, a cell with two copies of each chromosome. Errors in meiosis resulting in aneuploidy (an abnormal number of chromosomes) are the leading known cause of miscarriage and the most frequent genetic cause of developmental disabilities. In meiosis, DNA replication is followed by two rounds of cell division to produce four daughter cells, each with half the number of chromosomes as the original parent cell. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Biology
Biology is the scientific study of life and living organisms. It is a broad natural science that encompasses a wide range of fields and unifying principles that explain the structure, function, growth, History of life, origin, evolution, and distribution of life. Central to biology are five fundamental themes: the cell (biology), cell as the basic unit of life, genes and heredity as the basis of inheritance, evolution as the driver of biological diversity, energy transformation for sustaining life processes, and the maintenance of internal stability (homeostasis). Biology examines life across multiple biological organisation, levels of organization, from molecules and cells to organisms, populations, and ecosystems. Subdisciplines include molecular biology, physiology, ecology, evolutionary biology, developmental biology, and systematics, among others. Each of these fields applies a range of methods to investigate biological phenomena, including scientific method, observation, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
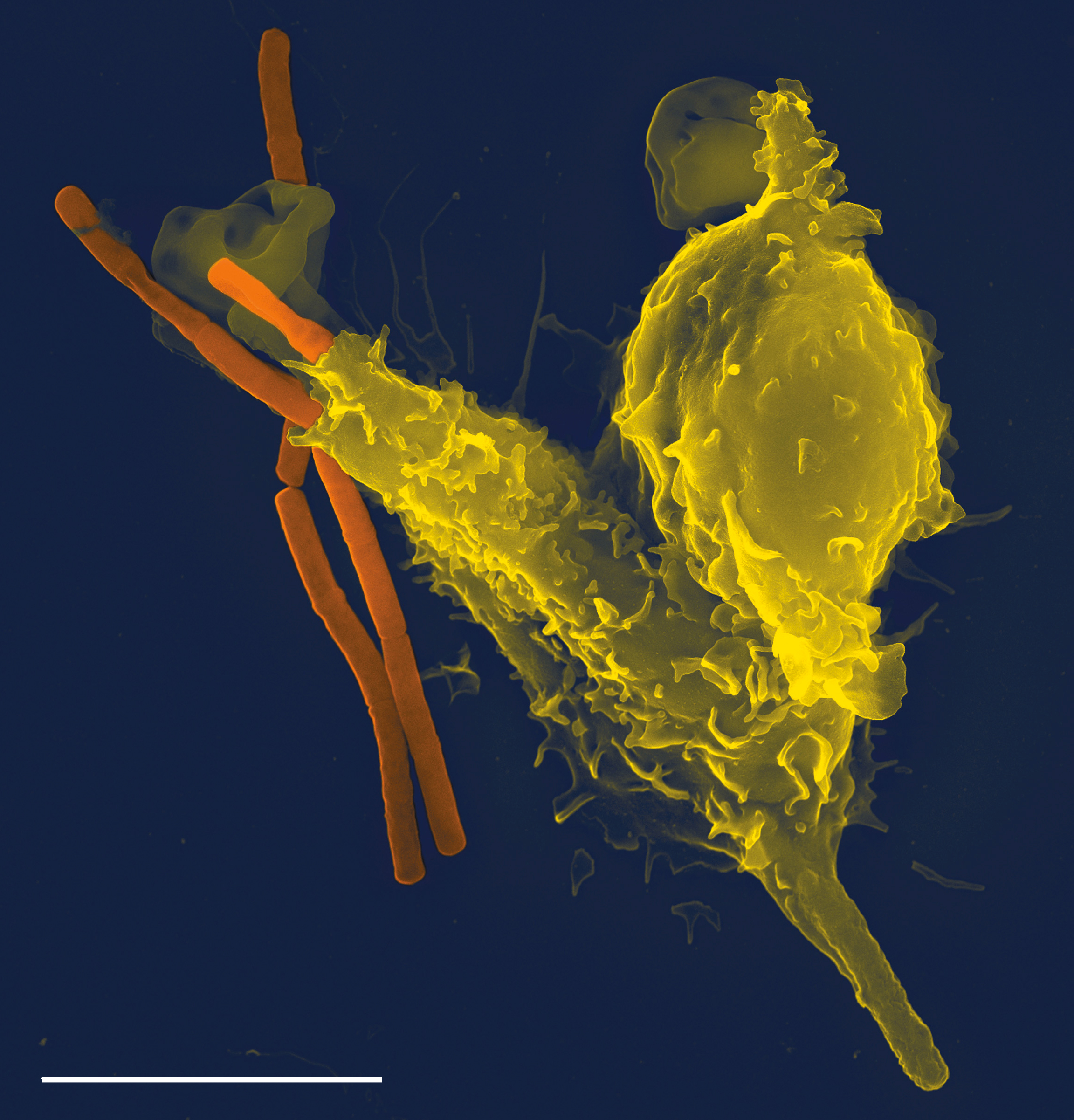
Immune System
The immune system is a network of biological systems that protects an organism from diseases. It detects and responds to a wide variety of pathogens, from viruses to bacteria, as well as Tumor immunology, cancer cells, Parasitic worm, parasitic worms, and also objects such as wood splinters, distinguishing them from the organism's own healthy biological tissue, tissue. Many species have two major subsystems of the immune system. The innate immune system provides a preconfigured response to broad groups of situations and stimuli. The adaptive immune system provides a tailored response to each stimulus by learning to recognize molecules it has previously encountered. Both use humoral immunity, molecules and cell-mediated immunity, cells to perform their functions. Nearly all organisms have some kind of immune system. Bacteria have a rudimentary immune system in the form of enzymes that protect against bacteriophage, viral infections. Other basic immune mechanisms evolved in ancien ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Darwin Hybrid Tulip Mutation 2014-05-01
Darwin may refer to: Common meanings * Charles Darwin (1809–1882), English naturalist and writer, best known as the originator of the theory of biological evolution by natural selection * Darwin, Northern Territory, a capital city in Australia, named after the naturalist Arts and entertainment * ''Darwin'' (1920 film), a German silent film * ''Darwin'' (2011 film), a documentary * ''Darwin'' (2015 film), a science fiction film by Alain Desrochers * Darwin (''seaQuest DSV''), a dolphin in the TV series ''seaQuest DSV'' * '' Darwin!'', a 1972 album by Banco del Mutuo Soccorso * '' Darwin: The Life of a Tormented Evolutionist'', a 1991 biography of Charles Darwin * Darwin (Marvel Comics), a Marvel Comics fictional superhero associated with the X-Men * Darwin Watterson, a character from the 2011 animated TV series ''The Amazing World of Gumball'' Computing * Darwin (ADL), an architecture description language * Darwin (operating system), the Unix base for Apple's iOS and macOS ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Phenotype
In genetics, the phenotype () is the set of observable characteristics or traits of an organism. The term covers the organism's morphology (physical form and structure), its developmental processes, its biochemical and physiological properties, and its behavior. An organism's phenotype results from two basic factors: the expression of an organism's genetic code (its genotype) and the influence of environmental factors. Both factors may interact, further affecting the phenotype. When two or more clearly different phenotypes exist in the same population of a species, the species is called polymorphic. A well-documented example of polymorphism is Labrador Retriever coloring; while the coat color depends on many genes, it is clearly seen in the environment as yellow, black, and brown. Richard Dawkins in 1978 and again in his 1982 book '' The Extended Phenotype'' suggested that one can regard bird nests and other built structures such as caddisfly larva cases and beaver dams ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Evolution
Evolution is the change in the heritable Phenotypic trait, characteristics of biological populations over successive generations. It occurs when evolutionary processes such as natural selection and genetic drift act on genetic variation, resulting in certain characteristics becoming more or less common within a population over successive generations. The process of evolution has given rise to biodiversity at every level of biological organisation. The scientific theory of evolution by natural selection was conceived independently by two British naturalists, Charles Darwin and Alfred Russel Wallace, in the mid-19th century as an explanation for why organisms are adapted to their physical and biological environments. The theory was first set out in detail in Darwin's book ''On the Origin of Species''. Evolution by natural selection is established by observable facts about living organisms: (1) more offspring are often produced than can possibly survive; (2) phenotypic variatio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Cancer
Cancer is a group of diseases involving Cell growth#Disorders, abnormal cell growth with the potential to Invasion (cancer), invade or Metastasis, spread to other parts of the body. These contrast with benign tumors, which do not spread. Possible Signs and symptoms of cancer, signs and symptoms include a lump, abnormal bleeding, prolonged cough, unexplained weight loss, and a change in defecation, bowel movements. While these symptoms may indicate cancer, they can also have other causes. List of cancer types, Over 100 types of cancers affect humans. Tobacco use is the cause of about 22% of cancer deaths. Another 10% are due to obesity, poor Diet (nutrition), diet, sedentary lifestyle, lack of physical activity or Alcohol abuse, excessive alcohol consumption. Other factors include certain infections, exposure to ionizing radiation, and environmental pollutants. infectious causes of cancer, Infection with specific viruses, bacteria and parasites is an environmental factor cau ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Single Chromosome Mutations
Single may refer to: Arts, entertainment, and media * Single (music), a song release Songs * Single (Natasha Bedingfield song), "Single" (Natasha Bedingfield song), 2004 * Single (New Kids on the Block and Ne-Yo song), "Single" (New Kids on the Block and Ne-Yo song), 2008 * Single (William Wei song), "Single" (William Wei song), 2016 * "Single", by Meghan Trainor from the album ''Meghan Trainor discography#Independent albums, Only 17'' * "Single", from the musical ''The Wedding Singer (musical)#Musical numbers, The Wedding Singer'' Film * Single (film), ''#Single'' (film), an Indian Telugu-language romantic comedy film Sports * Single (baseball), the most common type of base hit * Single (cricket), point in cricket * Single (football), Canadian football point * Single-speed bicycle Transportation * Single-cylinder engine, an internal combustion engine design with one cylinder, or a motorcycle using such engine * Single (locomotive), a steam locomotive with a single pair of driv ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Mobile Genetic Elements
Mobile genetic elements (MGEs), sometimes called selfish genetic elements, are a type of genetic material that can move around within a genome, or that can be transferred from one species or replicon to another. MGEs are found in all organisms. In humans, approximately 50% of the genome are thought to be MGEs. MGEs play a distinct role in evolution. Gene duplication events can also happen through the mechanism of MGEs. MGEs can also cause mutations in protein coding regions, which alters the protein functions. These mechanisms can also rearrange genes in the host genome generating variation. These mechanisms can increase fitness by gaining new or additional functions. An example of MGEs in evolutionary context are that virulence factors and antibiotic resistance genes of MGEs can be transported to share genetic code with neighboring bacteria. However, MGEs can also decrease fitness by introducing disease-causing alleles or mutations. The set of MGEs in an organism is called a mobil ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Genetic Variation
Genetic variation is the difference in DNA among individuals or the differences between populations among the same species. The multiple sources of genetic variation include mutation and genetic recombination. Mutations are the ultimate sources of genetic variation, but other mechanisms, such as genetic drift, contribute to it, as well. Among individuals within a population Genetic variation can be identified at many levels. Identifying genetic variation is possible from observations of phenotypic variation in either quantitative traits (traits that vary continuously and are coded for by many genes, e.g., leg length in dogs) or discrete traits (traits that fall into discrete categories and are coded for by one or a few genes, e.g., white, pink, or red petal color in certain flowers). Genetic variation can also be identified by examining variation at the level of enzymes using the process of protein electrophoresis. Polymorphic genes have more than one allele at each locu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
Natural Selection
Natural selection is the differential survival and reproduction of individuals due to differences in phenotype. It is a key mechanism of evolution, the change in the Heredity, heritable traits characteristic of a population over generations. Charles Darwin popularised the term "natural selection", contrasting it with selective breeding, artificial selection, which is intentional, whereas natural selection is not. Genetic diversity, Variation of traits, both Genotype, genotypic and phenotypic, exists within all populations of organisms. However, some traits are more likely to facilitate survival and reproductive success. Thus, these traits are passed the next generation. These traits can also become more Allele frequency, common within a population if the environment that favours these traits remains fixed. If new traits become more favoured due to changes in a specific Ecological niche, niche, microevolution occurs. If new traits become more favoured due to changes in the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |