Homing endonuclease on:
[Wikipedia]
[Google]
[Amazon]
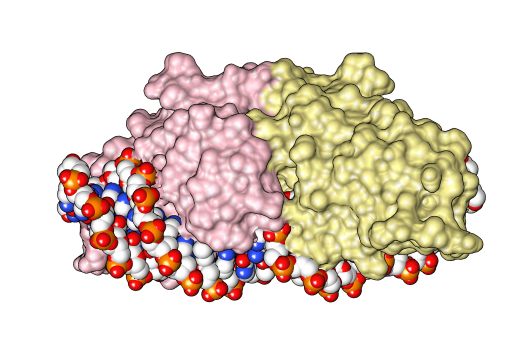 The homing endonucleases are a collection of
The homing endonucleases are a collection of
 The homing endonucleases are a collection of
The homing endonucleases are a collection of endonuclease
Endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (without regard to sequence), while many, typically called restriction endonucleases ...
s encoded either as freestanding gene
In biology, the word gene (from , ; "...Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a ba ...
s within intron
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e. a region inside a gene."The notion of the cistron .e., gene. ...
s, as fusions with host proteins, or as self-splicing inteins. They catalyze the hydrolysis of genomic DNA within the cells that synthesize them, but do so at very few, or even singular, locations. Repair of the hydrolyzed DNA by the host cell frequently results in the gene encoding the homing endonuclease having been copied into the cleavage site, hence the term 'homing' to describe the movement of these genes. Homing endonucleases can thereby transmit their genes horizontally within a host population, increasing their allele
An allele (, ; ; modern formation from Greek ἄλλος ''állos'', "other") is a variation of the same sequence of nucleotides at the same place on a long DNA molecule, as described in leading textbooks on genetics and evolution.
::"The chro ...
frequency at greater than Mendelian rates.
Origin and mechanism
Although the origin and function of homing endonucleases is still being researched, the most established hypothesis considers them asselfish genetic elements
Selfish genetic elements (historically also referred to as selfish genes, ultra-selfish genes, selfish DNA, parasitic DNA and genomic outlaws) are genetic segments that can enhance their own transmission at the expense of other genes in the genome, ...
, similar to transposon
A transposable element (TE, transposon, or jumping gene) is a nucleic acid sequence in DNA that can change its position within a genome, sometimes creating or reversing mutations and altering the cell's genetic identity and genome size. Transpo ...
s, because they facilitate the perpetuation of the genetic elements that encode them independent of providing a functional attribute to the host organism.
Homing endonuclease recognition sequences are long enough to occur randomly only with a very low probability (approximately once every ), and are normally found in one or very few instances per genome
In the fields of molecular biology and genetics, a genome is all the genetic information of an organism. It consists of nucleotide sequences of DNA (or RNA in RNA viruses). The nuclear genome includes protein-coding genes and non-coding ge ...
. Generally, owing to the homing mechanism, the gene encoding the endonuclease (the HEG, "homing endonuclease gene") is located within the recognition sequence which the enzyme cuts, thus interrupting the homing endonuclease recognition sequence and limiting DNA cutting only to sites that do not (yet) carry the HEG.
Prior to transmission, one allele
An allele (, ; ; modern formation from Greek ἄλλος ''állos'', "other") is a variation of the same sequence of nucleotides at the same place on a long DNA molecule, as described in leading textbooks on genetics and evolution.
::"The chro ...
carries the gene (HEG+) while the other does not (HEG−), and is therefore susceptible to being cut by the enzyme. Once the enzyme is synthesized, it breaks the chromosome in the HEG− allele, initiating a response from the cellular DNA repair
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA dam ...
system. The damage is repaired using recombination, taking the pattern of the opposite, undamaged DNA allele, HEG+, that contains the gene for the endonuclease. Thus, the gene is copied to the allele that initially did not have it and it is propagated through successive generations. This process is called "homing".
Nomenclature
Homing endonucleases are always indicated with a prefix that identifies their genomic origin, followed by a hyphen: "I-" for homing endonucleases encoded within an intron, "PI-" (for "protein insert") for those encoded within an intein. Some authors have proposed using the prefix "F-" ("freestanding") for viral enzymes and other natural enzymes not encoded by introns nor inteins, and "H-" ("hybrid") for enzymes synthesized in a laboratory. Next, a three-letter name is derived from the binominal name of the organism, taking one uppercase letter from thegenus
Genus ( plural genera ) is a taxonomic rank used in the biological classification of extant taxon, living and fossil organisms as well as Virus classification#ICTV classification, viruses. In the hierarchy of biological classification, genus com ...
name and two lowercase letters from the specific
Specific may refer to:
* Specificity (disambiguation)
* Specific, a cure or therapy for a specific illness
Law
* Specific deterrence, focussed on an individual
* Specific finding, intermediate verdict used by a jury in determining the fina ...
name. (Some mixing is usually done for hybrid enzymes.) Finally, a Roman numeral distinguishes different enzymes found in the same organism:
* PI-TliII () is the second-identified enzyme encoded by an intein found in the archaea
Archaea ( ; singular archaeon ) is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria (in the Archaebac ...
''Thermococcus litoralis
''Thermococcus litoralis'' (''T. litoralis'') is a species of Archaea that is found around deep-sea hydrothermal vents as well as shallow submarine thermal springs and oil wells. It is an anaerobic organotroph hyperthermophile that is between ...
''.
* H-DreI () is the first synthetic homing endonuclease, created in a laboratory from the enzymes I-DmoI () and I-CreI
I-''Cre''I is a homing endonuclease whose gene was first discovered in the chloroplast genome of ''Chlamydomonas reinhardtii'', a species of unicellular green algae. It is named for the facts that: it resides in an Intron; it was isolated from '' ...
(), taken respectively from '' Desulfurococcus mobilis'' and ''Chlamydomonas reinhardtii
''Chlamydomonas reinhardtii'' is a single-cell green alga about 10 micrometres in diameter that swims with two flagella. It has a cell wall made of hydroxyproline-rich glycoproteins, a large cup-shaped chloroplast, a large pyrenoid, and an ...
''.
Comparison to restriction enzymes
Homing endonucleases differ from Type II restriction enzymes in the several respects: * Whereas Type II restriction enzymes bind short, usually symmetric,recognition sequence
A recognition sequence is a DNA sequence to which a structural motif of a DNA-binding domain exhibits binding specificity. Recognition sequences are palindromes.
The transcription factor Sp1 for example, binds the sequences 5'-(G/T)GGGCGG(G/A)(G/ ...
s of 4 to 8 bp, homing endonucleases bind very long and in many cases asymmetric recognition sequences spanning 12 to 40 bp.
* Homing endonucleases are generally more tolerant of substitutions in the recognition sequence. Minor variations in the recognition sequence usually decrease the activity of homing endonucleases, but often do not completely abolish it as often occurs with restriction enzymes.
* Homing endonucleases share structural motif
In a polymer, chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a common Biomolecular structure#Tertiary structure, three-dimensional structure that appears in a variety of different, evolutionarily unrel ...
s that suggest there are four families, whereas it has not been possible to determine simply recognisable and distinguishable families of Type II restriction enzymes.
* Homing endonucleases act as monomer
In chemistry, a monomer ( ; ''mono-'', "one" + '' -mer'', "part") is a molecule that can react together with other monomer molecules to form a larger polymer chain or three-dimensional network in a process called polymerization.
Classification
Mo ...
s or homodimer
In biochemistry, a protein dimer is a macromolecular complex formed by two protein monomers, or single proteins, which are usually non-covalently bound. Many macromolecules, such as proteins or nucleic acids, form dimers. The word ''dimer'' ha ...
s, and often require associated proteins to regulate their activity or form ribonucleoprotein complexes, wherein RNA
Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and deoxyribonucleic acid ( DNA) are nucleic acids. Along with lipids, proteins, and carbohydra ...
is an integral component of the catalytic apparatus. Type II restriction enzymes can also function alone, as monomers or homodimers, or with additional protein subunit
In structural biology, a protein subunit is a polypeptide chain or single protein molecule that assembles (or "''coassembles''") with others to form a protein complex.
Large assemblies of proteins such as viruses often use a small number of ty ...
s, but the accessory subunits differ from those of the homing endonucleases. Thus, they can require restriction, modification, and specificity subunits for their action.
* Finally, homing endonucleases have a broader phylogenetic
In biology, phylogenetics (; from Greek φυλή/ φῦλον [] "tribe, clan, race", and wikt:γενετικός, γενετικός [] "origin, source, birth") is the study of the evolutionary history and relationships among or within groups o ...
distribution, occurring in all three biological domains—the archaea
Archaea ( ; singular archaeon ) is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria (in the Archaebac ...
, bacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among ...
and eukarya
Eukaryotes () are organisms whose cells have a nucleus. All animals, plants, fungi, and many unicellular organisms, are Eukaryotes. They belong to the group of organisms Eukaryota or Eukarya, which is one of the three domains of life. Bacte ...
. Type II restriction enzymes occur only in archaea, bacteria and certain viruses. Homing endonucleases are also expressed in all three compartments of the eukaryotic cell: nuclei, mitochondria
A mitochondrion (; ) is an organelle found in the Cell (biology), cells of most Eukaryotes, such as animals, plants and Fungus, fungi. Mitochondria have a double lipid bilayer, membrane structure and use aerobic respiration to generate adenosi ...
and chloroplast
A chloroplast () is a type of membrane-bound organelle known as a plastid that conducts photosynthesis mostly in plant and algal cells. The photosynthetic pigment chlorophyll captures the energy from sunlight, converts it, and stores it in ...
s. Open reading frames encoding homing endonucleases have been found in intron
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e. a region inside a gene."The notion of the cistron .e., gene. ...
s, inteins, and in freestanding form between genes, whereas genes encoding Type II restriction enzyme genes have been found only in freestanding form, almost always in close association with genes encoding cognate DNA modifying enzymes. Thus, while the Type II restriction enzymes and homing endonucleases share the function of cleaving double-stranded DNA, they appear to have evolved independently.
Structural families
Currently there are six known structural families. Their conservedstructural motif
In a polymer, chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a common Biomolecular structure#Tertiary structure, three-dimensional structure that appears in a variety of different, evolutionarily unrel ...
s are:
* LAGLIDADG: Every polypeptide has 1 or 2 LAGLIDADG motifs. The sequence LAGLIDADG is a conserved sequence of amino acid
Amino acids are organic compounds that contain both amino and carboxylic acid functional groups. Although hundreds of amino acids exist in nature, by far the most important are the alpha-amino acids, which comprise proteins. Only 22 alpha am ...
s where each letter is a code that identifies a specific residue. This sequence is directly involved in the DNA cutting process. Those enzymes that have only one motif work as homodimers, creating a saddle that interacts with the major groove
Major (commandant in certain jurisdictions) is a military rank of commissioned officer status, with corresponding ranks existing in many military forces throughout the world. When used unhyphenated and in conjunction with no other indicator ...
of each DNA half-site. The LAGLIDADG motifs contribute amino acid residues to both the protein-protein interface between protein domains or subunits, and to the enzyme's active sites. Enzymes that possess two motifs in a single protein chain act as monomers, creating the saddle in a similar way. The first structures to be determined of homing endonucleases (of PI-SceI and I-CreI, both reported in 1997) were both from the LAGLIDADG structural family., The following year, the first structure of a homing endonuclease (I-CreI) bound to its DNA target site was also reported.
* GIY-YIG: These have only one GIY-YIG motif, in the N-terminal
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the ami ...
region, that interacts with the DNA in the cutting site. The prototypic enzyme of this family is I-TevI which acts as a monomer. Separate structural studies have been reported of the DNA-binding and catalytic domains of I-TevI, the former bound to its DNA target and the latter in the absence of DNA.,
* His-Cys box (): These enzymes possess a region of 30 amino acids that includes 5 conserved residues: two histidine
Histidine (symbol His or H) is an essential amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated –NH3+ form under biological conditions), a carboxylic acid group (which is in the de ...
s and three cystein
Cysteine (symbol Cys or C; ) is a semiessential proteinogenic amino acid with the formula . The thiol side chain in cysteine often participates in enzymatic reactions as a nucleophile.
When present as a deprotonated catalytic residue, someti ...
s. They co-ordinate the metal cation needed for catalysis. I-PpoI is the best characterized enzyme of this family and acts as a homodimer. Its structure was reported in 1998. It is possibly related to the H-N-H family, as they share common features.
* H-N-H: (): These have a consensus sequence of approximately 30 amino acids. It includes two pairs of conserved histidine
Histidine (symbol His or H) is an essential amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated –NH3+ form under biological conditions), a carboxylic acid group (which is in the de ...
s and one asparagine
Asparagine (symbol Asn or N) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α-amino group (which is in the protonated −NH form under biological conditions), an α-carboxylic acid group (which is in the depro ...
that create a zinc finger
A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) in order to stabilize the fold. It was originally coined to describe the finger-like appearance of a hypothesized struct ...
domain. I-HmuI () is the best characterized enzyme of this family, and acts as a monomer. Its structure was reported in 2004 ().
* PD-(D/E)xK (): These enzymes contain a canonical nuclease catalytic domain typically found in type II restriction endonucleases. The best characterized enzyme in this family, I-Ssp6803I (), acts as a tetramer. Its structure was reported in 2007 (). The overall fold is conserved in many endonuclease families, all of which belong to the PD-(D/E)xK superfamily.
* Vsr-like/EDxHD (DUF559, ): These enzymes were discovered in the Global Ocean Sampling Metagenomic Database and first described in 2009. The term 'Vsr-like' refers to the presence of a C-terminal nuclease domain that displays recognizable homology to bacterial Very short patch repair
Very short patch (VSP) repair is a DNA repair system that removes GT mismatches created by the deamination of 5-methylcytosine to thymine. This system exists because the glycosylases which normally target deaminated bases cannot target thymine (it ...
(Vsr) endonucleases. The structure has been solved in 2011, confirming the Vsr homology. Is considered part of the PD-(D/E)xk superfamily.
Domain architecture
The yeast homing endonuclease PI-Sce is a LAGLIDADG-type endonuclease encoded as an intein that splices itself out of another protein (). The high-resolution structure reveals two domains: an endonucleolytic centre resembling theC-terminal
The C-terminus (also known as the carboxyl-terminus, carboxy-terminus, C-terminal tail, C-terminal end, or COOH-terminus) is the end of an amino acid chain (protein or polypeptide), terminated by a free carboxyl group (-COOH). When the protein is ...
domain of Hedgehog proteins, and a Hint domain
Hint and similar may refer to:
* Hint (musician), musician Jonathan James from Sussex, England
* Hint (SQL), a feature of the SQL computer language
* Hint Water, a beverage company from San Francisco, California
* Aadu Hint (1910–1989), Estonian ...
(Hedgehog/Intein) containing the protein-splicing active site
In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate (binding site) a ...
.
See also
* REBASE, a comprehensive restriction enzyme database fromNew England Biolabs
New England Biolabs (NEB) produces and supplies recombinant and native enzyme reagents for the life science research, as well as providing products and services supporting genome editing, synthetic biology and next-generation sequencing. NEB al ...
with links to related literature.
* List of homing endonuclease cutting sites
* I-CreI homing endonuclease
* Meganucleases
* Restriction enzyme
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class o ...
* Intron
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e. a region inside a gene."The notion of the cistron .e., gene. ...
s and inteins
* Intragenomic conflict: Homing endonuclease genes
* Transposon
A transposable element (TE, transposon, or jumping gene) is a nucleic acid sequence in DNA that can change its position within a genome, sometimes creating or reversing mutations and altering the cell's genetic identity and genome size. Transpo ...
References
External links
* * {{DEFAULTSORT:Homing Endonuclease Protein domains Molecular biology Biotechnology Restriction enzymes