|
Recognition Sequence
A recognition sequence is a DNA sequence to which a structural motif of a DNA-binding domain exhibits binding specificity. Recognition sequences are palindromes. The transcription factor Sp1 for example, binds the sequences 5'-(G/T)GGGCGG(G/A)(G/A)(C/T)-3', where (G/T) indicates that the domain will bind a guanine or thymine at this position. The restriction endonuclease PstI recognizes, binds, and cleaves the sequence 5'-CTGCAG-3'. A recognition sequence is different from a recognition site. A given recognition sequence can occur one or more times, or not at all, on a specific DNA fragment. A recognition site is specified by the position of the site. For example, there are two PstI recognition sites in the following DNA sequence fragment, starting at base 9 and 31 respectively. A recognition sequence is a specific sequence, usually very short (less than 10 bases). Depending on the degree of specificity of the protein, a DNA-binding protein can bind to more than one specific sequ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Sequence
DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery. Knowledge of DNA sequences has become indispensable for basic biological research, DNA Genographic Projects and in numerous applied fields such as medical diagnosis, biotechnology, forensic biology, virology and biological systematics. Comparing healthy and mutated DNA sequences can diagnose different diseases including various cancers, characterize antibody repertoire, and can be used to guide patient treatment. Having a quick way to sequence DNA allows for faster and more individualized medical care to be administered, and for more organisms to be identified and cataloged. The rapid speed of sequencing attained with modern D ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Structural Motif
In a polymer, chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a common Biomolecular structure#Tertiary structure, three-dimensional structure that appears in a variety of different, evolutionarily unrelated molecules. A structural motif does not have to be associated with a sequence motif; it can be represented by different and completely unrelated sequences in different proteins or RNA. In nucleic acids Depending upon the sequence and other conditions, nucleic acids can form a variety of structural motifs which is thought to have biological significance. ;Stem-loop: Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded DNA or, more commonly, in RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions, base-pair to form a double helix that ends in an unpaired loop. The re ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
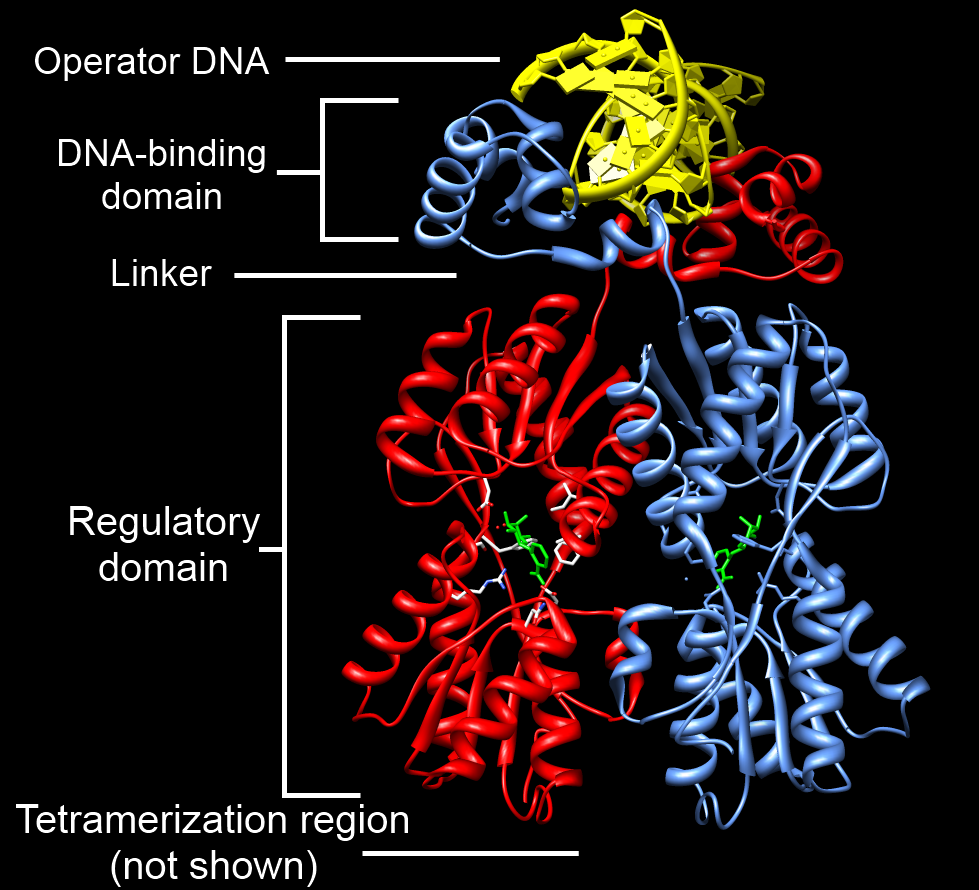
DNA-binding Domain
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence (a recognition sequence) or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure. Function One or more DNA-binding domains are often part of a larger protein consisting of further protein domains with differing function. The extra domains often regulate the activity of the DNA-binding domain. The function of DNA binding is either structural or involves transcription regulation, with the two roles sometimes overlapping. DNA-binding domains with functions involving DNA structure have biological roles in DNA replication, repair, storage, and modification, such as methylation. Many proteins involved in the regulation of gene expression contain DNA-binding domains. For example, proteins that regulate transcription by ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Palindromic Sequence
A palindromic sequence is a nucleic acid sequence in a double-stranded DNA or RNA molecule whereby reading in a certain direction (e.g. 5' to 3') on one strand is identical to the sequence in the same direction (e.g. 5' to 3') on the complementary strand. This definition of palindrome thus depends on complementary strands being palindromic of each other. The meaning of palindrome in the context of genetics is slightly different from the definition used for words and sentences. Since a double helix is formed by two paired antiparallel strands of nucleotides that run in opposite directions, and the nucleotides always pair in the same way (adenine (A) with thymine (T) in DNA or uracil (U) in RNA; cytosine (C) with guanine (G)), a (single-stranded) nucleotide sequence is said to be a palindrome if it is equal to its reverse complement. For example, the DNA sequence ACCTAGGT is palindromic with its nucleotide-by-nucleotide complement TGGATCCA because reversing the order of the nuc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sp1 (biology)
Transcription factor Sp1, also known as specificity protein 1* is a protein that in humans is encoded by the SP1 gene. Function The protein encoded by this gene is a zinc finger transcription factor that binds to GC-rich motifs of many promoters. The encoded protein is involved in many cellular processes, including cell differentiation, cell growth, apoptosis, immune responses, response to DNA damage, and chromatin remodeling. Post-translational modifications Post-translational modification (PTM) is the covalent and generally enzymatic modification of proteins following protein biosynthesis. This process occurs in the endoplasmic reticulum and the golgi apparatus. Proteins are synthesized by ribosom ... such as Protein phosphorylation, phosphorylation, acetylation, O-GlcNAc, ''O''-GlcNAcylation, and proteolytic processing significantly affect the activity of this protein, which can be an activator or a repressor. In the SV40 virus, Sp1 binds to the GC boxes in the regula ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Restriction Endonuclease
A restriction enzyme, restriction endonuclease, REase, ENase or'' restrictase '' is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone (i.e. each strand) of the DNA double helix. These enzymes are found in bacteria and archaea and provide a defense mechanism against invading viruses. Inside a prokaryote, the restriction enzymes selectively cut up ''foreign'' DNA in a process called ''restriction digestion''; meanwhile, host DNA is protected by a modification enzyme (a methyltransferase) that modifi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PstI
''Pst''I is a type II restriction endonuclease isolated from the Gram negative species, '' Providencia stuartii''. Function ''Pst''I cleaves DNA at the recognition sequence 5′-CTGCA/G-3′ generating fragments with 3′-cohesive termini. This cleavage yields sticky ends 4 base pairs long. PstI is catalytically active as a dimer. The two subunits are related by a 2-fold symmetry axis which in the complex with the substrate coincides with the dyad axis of the recognition sequence. It has a molecular weight of 69,500 and contains 54 positive and 41 negatively charged residues. The PstI restriction/modification (R/M) system has two components: a restriction enzyme that cleaves foreign DNA, and a methyltransferase which protect native DNA strands by methylation of the adenine base inside the recognition sequence. The combination of both provide is a defense mechanism against invading viruses. The methyltransferase and endonuclease are encoded as two separate proteins and act in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription Factor
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The function of TFs is to regulate—turn on and off—genes in order to make sure that they are expressed in the desired cells at the right time and in the right amount throughout the life of the cell and the organism. Groups of TFs function in a coordinated fashion to direct cell division, cell growth, and cell death throughout life; cell migration and organization (body plan) during embryonic development; and intermittently in response to signals from outside the cell, such as a hormone. There are up to 1600 TFs in the human genome. Transcription factors are members of the proteome as well as regulome. TFs work alone or with other proteins in a complex, by promoting (as an activator), or blocking (as a repressor) the recruitment of RNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetics
Genetics is the study of genes, genetic variation, and heredity in organisms.Hartl D, Jones E (2005) It is an important branch in biology because heredity is vital to organisms' evolution. Gregor Mendel, a Moravian Augustinian friar working in the 19th century in Brno, was the first to study genetics scientifically. Mendel studied "trait inheritance", patterns in the way traits are handed down from parents to offspring over time. He observed that organisms (pea plants) inherit traits by way of discrete "units of inheritance". This term, still used today, is a somewhat ambiguous definition of what is referred to as a gene. Trait inheritance and molecular inheritance mechanisms of genes are still primary principles of genetics in the 21st century, but modern genetics has expanded to study the function and behavior of genes. Gene structure and function, variation, and distribution are studied within the context of the cell, the organism (e.g. dominance), and within the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genome Editing
Genome editing, or genome engineering, or gene editing, is a type of genetic engineering in which DNA is inserted, deleted, modified or replaced in the genome of a living organism. Unlike early genetic engineering techniques that randomly inserts genetic material into a host genome, genome editing targets the insertions to site-specific locations. The basic mechanism involved in genetic manipulations through programmable nucleases is the recognition of target genomic loci and binding of effector DNA-binding domain (DBD), double-strand breaks (DSBs) in target DNA by the restriction endonucleases ( FokI and Cas), and the repair of DSBs through homology-directed recombination (HDR) or non-homologous end joining (NHEJ). History Genome editing was pioneered in the 1990s, before the advent of the common current nuclease-based gene editing platforms, however, its use was limited by low efficiencies of editing. Genome editing with engineered nucleases, i.e. all three major classes of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |