|
Structural Motif
In a chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a common three-dimensional structure that appears in a variety of different, evolutionarily unrelated molecules. A structural motif does not have to be associated with a sequence motif; it can be represented by different and completely unrelated sequences in different proteins or RNA. In nucleic acids Depending upon the sequence and other conditions, nucleic acids can form a variety of structural motifs which is thought to have biological significance. ;Stem-loop: Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded DNA or, more commonly, in RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions, base-pair to form a double helix that ends in an unpaired loop. The resulting structure is a key building block of many ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymer
A polymer (; Greek ''poly-'', "many" + '' -mer'', "part") is a substance or material consisting of very large molecules called macromolecules, composed of many repeating subunits. Due to their broad spectrum of properties, both synthetic and natural polymers play essential and ubiquitous roles in everyday life. Polymers range from familiar synthetic plastics such as polystyrene to natural biopolymers such as DNA and proteins that are fundamental to biological structure and function. Polymers, both natural and synthetic, are created via polymerization of many small molecules, known as monomers. Their consequently large molecular mass, relative to small molecule compounds, produces unique physical properties including toughness, high elasticity, viscoelasticity, and a tendency to form amorphous and semicrystalline structures rather than crystals. The term "polymer" derives from the Greek word πολύς (''polus'', meaning "many, much") and μέρος (''meros'', mean ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Complementarity (molecular Biology)
In molecular biology, complementarity describes a relationship between two structures each following the lock-and-key principle. In nature complementarity is the base principle of DNA replication and transcription as it is a property shared between two DNA or RNA sequences, such that when they are aligned antiparallel to each other, the nucleotide bases at each position in the sequences will be complementary, much like looking in the mirror and seeing the reverse of things. This complementary base pairing allows cells to copy information from one generation to another and even find and repair damage to the information stored in the sequences. The degree of complementarity between two nucleic acid strands may vary, from complete complementarity (each nucleotide is across from its opposite) to no complementarity (each nucleotide is not across from its opposite) and determines the stability of the sequences to be together. Furthermore, various DNA repair functions as well as ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nest (protein Structural Motif)
The Nest is a type of protein structural motif. It is a small recurring anion-binding feature of both proteins and peptides. Each consists of the main chain atoms of three consecutive amino acid residues. The main chain NH groups bind the anions while the side chain atoms are often not involved. Proline residues lack NH groups so are rare in nests. About one in 12 of amino acid residues in proteins, on average, belongs to a nest. Nest conformations The conformation of a nest is such that the NH groups of the first and third amino acid residues are liable to be hydrogen bonded to a negatively charged, or partially negatively charged, atom, often an oxygen atom. The NH of the second residue may also be hydrogen bonded to the same atom but usually points somewhat away. These main chain atoms form a concavity called a nest into which an anionic atom fits. Such anionic atoms are sometimes called eggs and more than one egg may occur bound to a nest. The oxyanion hole of the intest ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Helix-turn-helix
Helix-turn-helix is a DNA-binding protein (DBP). The helix-turn-helix (HTH) is a major structural motif capable of binding DNA. Each monomer incorporates two α helices, joined by a short strand of amino acids, that bind to the major groove of DNA. The HTH motif occurs in many proteins that regulate gene expression. It should not be confused with the helix–loop–helix motif. Discovery The discovery of the helix-turn-helix motif was based on similarities between several genes encoding transcription regulatory proteins from bacteriophage lambda and ''Escherichia coli'': Cro, CAP, and λ repressor, which were found to share a common 20–25 amino acid sequence that facilitates DNA recognition. Function The helix-turn-helix motif is a DNA-binding motif. The recognition and binding to DNA by helix-turn-helix proteins is done by the two α helices, one occupying the N-terminal end of the motif, the other at the C-terminus. In most cases, such as in the Cro repressor, the s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Zinc Finger
A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) in order to stabilize the fold. It was originally coined to describe the finger-like appearance of a hypothesized structure from the African clawed frog (''Xenopus laevis'') transcription factor IIIA. However, it has been found to encompass a wide variety of differing protein structures in eukaryotic cells. '' Xenopus laevis'' TFIIIA was originally demonstrated to contain zinc and require the metal for function in 1983, the first such reported zinc requirement for a gene regulatory protein followed soon thereafter by the Krüppel factor in ''Drosophila''. It often appears as a metal-binding domain in multi-domain proteins. Proteins that contain zinc fingers (zinc finger proteins) are classified into several different structural families. Unlike many other clearly defined supersecondary structures such as Greek keys or β hairpins, there are a number of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helix
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand-helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence. The alpha helix is also called a classic Pauling–Corey–Branson α-helix. The name 3.613-helix is also used for this type of helix, denoting the average number of residues per helical turn, with 13 atoms being involved in the ring formed by the hydrogen bond. Among types of local structure in proteins, the α-helix is the most extreme and the most predictable from sequence, as well as the most prevalent. Discovery In the early 1930s, William Astbury showed that there were drastic changes in the X-ray fiber diffraction of moist wool or hair fibers upon significant stretching. The data suggested that the unstretched fibers had a coiled molecular structure with a characteristic repeat of ≈. Astbu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Omega Loop
The omega loop is a non-regular protein structural motif, consisting of a loop of six or more amino acid residues and any amino acid sequence. The defining characteristic is that residues that make up the beginning and end of the loop are close together in space with no intervening lengths of regular secondary structural motifs. It is named after its shape, which resembles the upper-case Greek letter Omega (Ω). Structure Omega loops, being non-regular, non-repeating secondary structural units, have a variety of three-dimensional shapes. Omega loop shapes are analyzed to identify recurring patterns in dihedral angles and overall loop shape to help identify potential roles in protein folding and function. Since loops are almost always at the protein surface, it is often assumed that these structures are flexible; however, different omega loops exhibit ranges of flexibility across different time scales of protein motion and have been identified as playing a role in the folding of som ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta Sheet
The beta sheet, (β-sheet) (also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, notably Alzheimer's disease. History The first β-sheet structure was proposed by William Astbury in the 1930s. He proposed the idea of hydrogen bonding between the peptide bonds of parallel or antiparallel extended β-strands. However, Astbury did not have the necessary data on the bond geometry of the amino acids in order to build accurate models, especially since he did not then know that the peptide bond was planar. A refined version was ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antiparallel (biochemistry)
In biochemistry, two biopolymers are antiparallel if they run parallel to each other but with opposite directionality (alignments). An example is the two complementary strands of a DNA double helix, which run in opposite directions alongside each other. Nucleic acids Nucleic acid molecules have a phosphoryl (5') end and a hydroxyl (3') end. This notation follows from organic chemistry nomenclature, and can be used to define the movement of enzymes such as DNA polymerases relative to the DNA strand in a non-arbitrary manner. G-quadruplexes G-quadruplexes, also known as G4 DNA are secondary structures found in nucleic acids that are rich in guanine. These structures are normally located at the telomeres (the ends of the chromosomes). The G-quadruplex can either be parallel or antiparallel depending on the loop configuration, which is a component of the structure. If all the DNA strands run in the same direction, it is termed to be a parallel quadruplex, and is known as a s ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
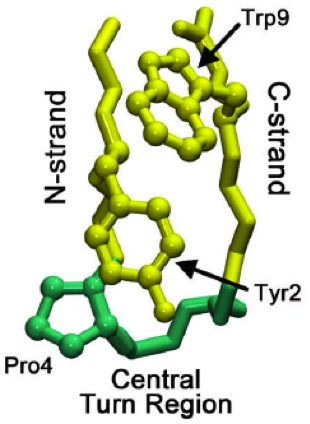
Beta Hairpin
The beta hairpin (sometimes also called beta-ribbon or beta-beta unit) is a simple protein structural motif involving two beta strands that look like a hairpin. The motif consists of two strands that are adjacent in primary structure, oriented in an antiparallel direction (the N-terminus of one sheet is adjacent to the C-terminus of the next), and linked by a short loop of two to five amino acids. Beta hairpins can occur in isolation or as part of a series of hydrogen bonded strands that collectively comprise a beta sheet. Researchers such as Francisco Blanco ''et al.'' have used protein NMR to show that beta-hairpins can be formed from isolated short peptides in aqueous solution, suggesting that hairpins could form nucleation sites for protein folding. Classification Beta hairpins were originally categorized solely by the number of amino acid residues in their loop sequences, such that they were named one-residue, two-residue, etc. This system, however, is somewhat ambiguous ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Tandem Repeats
An array of protein tandem repeats is defined as several (at least two) adjacent copies having the same or similar sequence motifs. These periodic sequences are generated by internal duplications in both coding and non-coding genomic sequences. Repetitive units of protein tandem repeats are considerably diverse, ranging from the repetition of a single amino acid to domains of 100 or more residues. "Repeats" in proteins In proteins, a "repeat" is any sequence block that returns more than one time in the sequence, either in an identical or a highly similar form. The degree of similarity can be highly variable, with some repeats maintaining only a few conserved amino acid positions and a characteristic length. Highly degenerate repeats can be very difficult to detect from sequence alone. Structural similarity can help to identify repetitive patterns in sequence. Structure Repetitiveness does not in itself indicate anything about the structure of the protein. As a "rule of thu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |




_structure.png)