|
Transcription-translation Coupling
Transcription-translation coupling is a mechanism of regulation of gene expression, gene expression regulation in which synthesis of an mRNA (transcription (biology), transcription) is affected by its concurrent decoding (translation (biology), translation). In prokaryotes, mRNAs are translated while they are transcribed. This allows communication between RNA polymerase, the multisubunit enzyme that catalyzes transcription, and the ribosome, which catalyzes translation. Coupling involves both direct physical interactions between RNA polymerase and the ribosome ("expressome" complexes), as well as ribosome-induced changes to the structure and accessibility of the intervening mRNA that affect transcription ("attenuation" and "polarity"). Significance Bacteria depend on transcription-translation coupling for Chromosome instability#Genome integrity, genome integrity, Transcription (biology)#Termination, termination of transcription and control of Messenger RNA#Degradation, mRNA stabilit ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Regulation Of Gene Expression
Regulation of gene expression, or gene regulation, includes a wide range of mechanisms that are used by cells to increase or decrease the production of specific gene products (protein or RNA). Sophisticated programs of gene expression are widely observed in biology, for example to trigger developmental pathways, respond to environmental stimuli, or adapt to new food sources. Virtually any step of gene expression can be modulated, from Transcriptional regulation, transcriptional initiation, to RNA processing, and to the post-translational modification of a protein. Often, one gene regulator controls another, and so on, in a gene regulatory network. Gene regulation is essential for viruses, prokaryotes and eukaryotes as it increases the versatility and adaptability of an organism by allowing the cell to express protein when needed. Although as early as 1951, Barbara McClintock showed interaction between two genetic loci, Activator (''Ac'') and Dissociator (''Ds''), in the color f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Escherichia Coli
''Escherichia coli'' (),Wells, J. C. (2000) Longman Pronunciation Dictionary. Harlow ngland Pearson Education Ltd. also known as ''E. coli'' (), is a Gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus ''Escherichia'' that is commonly found in the lower intestine of warm-blooded organisms. Most ''E. coli'' strains are harmless, but some serotypes ( EPEC, ETEC etc.) can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. Most strains do not cause disease in humans and are part of the normal microbiota of the gut; such strains are harmless or even beneficial to humans (although these strains tend to be less studied than the pathogenic ones). For example, some strains of ''E. coli'' benefit their hosts by producing vitamin K2 or by preventing the colonization of the intestine by pathogenic bacteria. These mutually beneficial relationships between ''E. col ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Intrinsic Termination
Intrinsic, or rho-independent termination, is a process in prokaryotes to signal the end of transcription and release the newly constructed RNA molecule. In prokaryotes such as E. coli, transcription is terminated either by a rho-dependent process or rho-independent process. In the Rho-dependent process, the rho-protein locates and binds the signal sequence in the mRNA and signals for cleavage. Contrarily, intrinsic termination does not require a special protein to signal for termination and is controlled by the specific sequences of RNA. When the termination process begins, the transcribed mRNA forms a stable secondary structure hairpin loop, also known as a Stem-loop. This RNA hairpin is followed by multiple uracil nucleotides. The bonds between uracil and adenine are very weak. A protein bound to RNA polymerase (nusA) binds to the stem-loop structure tightly enough to cause the polymerase to temporarily stall. This pausing of the polymerase coincides with transcription of the po ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Operon
In genetics, an operon is a functioning unit of DNA containing a cluster of genes under the control of a single promoter. The genes are transcribed together into an mRNA strand and either translated together in the cytoplasm, or undergo splicing to create monocistronic mRNAs that are translated separately, i.e. several strands of mRNA that each encode a single gene product. The result of this is that the genes contained in the operon are either expressed together or not at all. Several genes must be ''co-transcribed'' to define an operon. Originally, operons were thought to exist solely in prokaryotes (which includes organelles like plastids that are derived from bacteria), but since the discovery of the first operons in eukaryotes in the early 1990s, more evidence has arisen to suggest they are more common than previously assumed. In general, expression of prokaryotic operons leads to the generation of polycistronic mRNAs, while eukaryotic operons lead to monocistronic mRNA ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nonsense Mutation
In genetics, a nonsense mutation is a point mutation in a sequence of DNA that results in a premature stop codon, or a ''nonsense codon'' in the transcribed mRNA, and in leading to a truncated, incomplete, and usually nonfunctional protein product. The functional effect of a nonsense mutation depends on the location of the stop codon within the coding DNA. For example, the effect of a nonsense mutation depends on the proximity of the nonsense mutation to the original stop codon, and the degree to which functional subdomains of the protein are affected. As nonsense mutations leads to premature termination of polypeptide chains; they are also called chain termination mutations. Missense mutations differ from nonsense mutations since they are point mutations that exhibit a single nucleotide change to cause substitution of a different amino acid. A nonsense mutation also differs from a nonstop mutation, which is a point mutation that removes a stop codon. About 10% of patients facin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Repressor
In molecular genetics, a repressor is a DNA- or RNA-binding protein that inhibits the expression of one or more genes by binding to the operator or associated silencers. A DNA-binding repressor blocks the attachment of RNA polymerase to the promoter, thus preventing transcription of the genes into messenger RNA. An RNA-binding repressor binds to the mRNA and prevents translation of the mRNA into protein. This blocking or reducing of expression is called repression. Function If an inducer, a molecule that initiates the gene expression, is present, then it can interact with the repressor protein and detach it from the operator. RNA polymerase then can transcribe the message (expressing the gene). A co-repressor is a molecule that can bind to the repressor and make it bind to the operator tightly, which decreases transcription. A repressor that binds with a co-repressor is termed an ''aporepressor'' or ''inactive repressor''. One type of aporepressor is the trp repressor, a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Lambda Phage
''Enterobacteria phage λ'' (lambda phage, coliphage λ, officially ''Escherichia virus Lambda'') is a bacterial virus, or bacteriophage, that infects the bacterial species ''Escherichia coli'' (''E. coli''). It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell. The phage particle consists of a head (also known as a capsid), a tail, and tail fibers (see image of virus below). The head contains the phage's double-strand linear DNA genome. During infection, the phage particle recognizes and binds to its host, ''E. coli'', causing DNA in the head of the phage to be ejected through the tail into the ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Open Reading Frame
In molecular biology, open reading frames (ORFs) are defined as spans of DNA sequence between the start and stop codons. Usually, this is considered within a studied region of a prokaryotic DNA sequence, where only one of the six possible reading frames will be "open" (the "reading", however, refers to the RNA produced by transcription of the DNA and its subsequent interaction with the ribosome in translation). Such an ORF may contain a start codon (usually AUG in terms of RNA) and by definition cannot extend beyond a stop codon (usually UAA, UAG or UGA in RNA). That start codon (not necessarily the first) indicates where translation may start. The transcription termination site is located after the ORF, beyond the translation stop codon. If transcription were to cease before the stop codon, an incomplete protein would be made during translation. In eukaryotic genes with multiple exons, introns are removed and exons are then joined together after transcription to yield the final ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Operons
In genetics, an operon is a functioning unit of DNA containing a cluster of genes under the control of a single promoter. The genes are transcribed together into an mRNA strand and either translated together in the cytoplasm, or undergo splicing to create monocistronic mRNAs that are translated separately, i.e. several strands of mRNA that each encode a single gene product. The result of this is that the genes contained in the operon are either expressed together or not at all. Several genes must be ''co-transcribed'' to define an operon. Originally, operons were thought to exist solely in prokaryotes (which includes organelles like plastids that are derived from bacteria), but since the discovery of the first operons in eukaryotes in the early 1990s, more evidence has arisen to suggest they are more common than previously assumed. In general, expression of prokaryotic operons leads to the generation of polycistronic mRNAs, while eukaryotic operons lead to monocistronic mRNAs ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacillus Subtilis
''Bacillus subtilis'', known also as the hay bacillus or grass bacillus, is a Gram-positive, catalase-positive bacterium, found in soil and the gastrointestinal tract of ruminants, humans and marine sponges. As a member of the genus ''Bacillus'', ''B. subtilis'' is rod-shaped, and can form a tough, protective endospore, allowing it to tolerate extreme environmental conditions. ''B. subtilis'' has historically been classified as an obligate aerobe, though evidence exists that it is a facultative anaerobe. ''B. subtilis'' is considered the best studied Gram-positive bacterium and a model organism to study bacterial chromosome replication and cell differentiation. It is one of the bacterial champions in secreted enzyme production and used on an industrial scale by biotechnology companies. Description ''Bacillus subtilis'' is a Gram-positive bacterium, rod-shaped and catalase-positive. It was originally named ''Vibrio subtilis'' by Christian Gottfried Ehrenberg, and renamed ''B ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
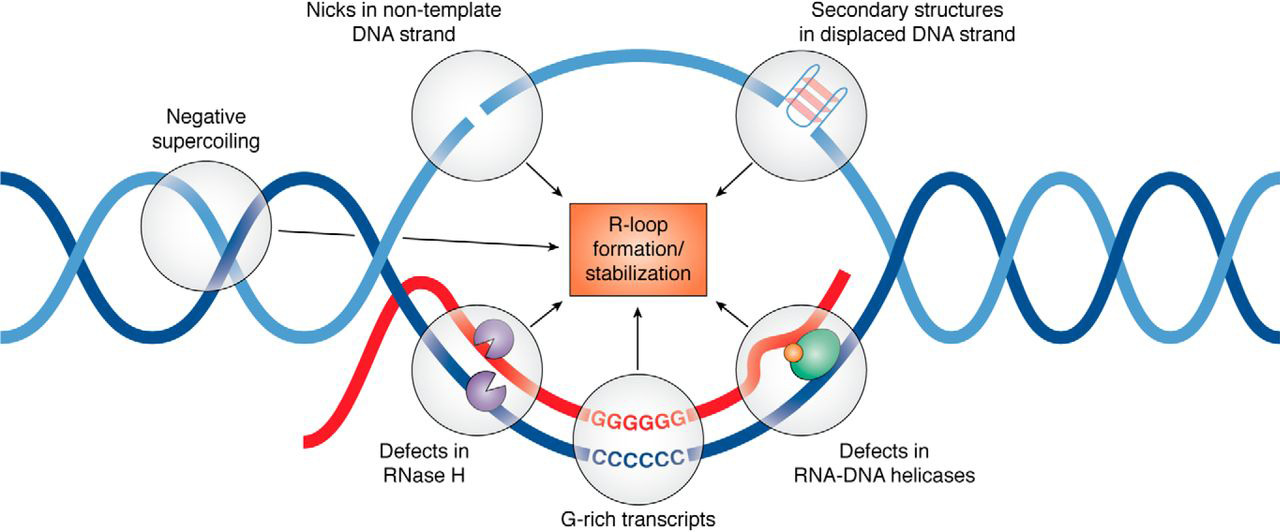
R-loop
An R-loop is a three-stranded nucleic acid structure, composed of a DNA:RNA hybrid and the associated non-template single-stranded DNA. R-loops may be formed in a variety of circumstances, and may be tolerated or cleared by cellular components. The term "R-loop" was given to reflect the similarity of these structures to D-loops; the "R" in this case represents the involvement of an RNA moiety. In the laboratory, R-loops may also be created by the hybridization of mature mRNA with double-stranded DNA under conditions favoring the formation of a DNA-RNA hybrid; in this case, the intron regions (which have been spliced out of the mRNA) form single-stranded DNA loops, as they cannot hybridize with complementary sequence in the mRNA. History R-looping was first described in 1976. Independent R-looping studies from the laboratories of Richard J. Roberts and Phillip A. Sharp showed that protein coding adenovirus genes contained DNA sequences that were not present in the mature mRNA. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcription (biology)
Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins are said to produce messenger RNA (mRNA). Other segments of DNA are copied into RNA molecules called non-coding RNAs (ncRNAs). mRNA comprises only 1–3% of total RNA samples. Less than 2% of the human genome can be transcribed into mRNA ( Human genome#Coding vs. noncoding DNA), while at least 80% of mammalian genomic DNA can be actively transcribed (in one or more types of cells), with the majority of this 80% considered to be ncRNA. Both DNA and RNA are nucleic acids, which use base pairs of nucleotides as a complementary language. During transcription, a DNA sequence is read by an RNA polymerase, which produces a complementary, antiparallel RNA strand called a primary transcript. Transcription proceeds in the following general steps: # RNA polymerase, together with one or more general transcription factors, binds to promoter ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |