|
YbhL Leader
The YbhL leader is a putative structured RNA element that is found upstream of the uncharacterized YbhL membrane protein in alpha-proteobacteria. Other non-coding RNAs uncovered in the same analysis include: speF, suhB ''suhB'', also known as ''mmgR'' (makes more granules regulator), is a non-coding RNA found multiple times in the ''Agrobacterium tumefaciens'' genome and related alpha-proteobacteria. Other non-coding RNAs uncovered in the same analysis include ..., metA and serC. References External links * Cis-regulatory RNA elements {{molecular-cell-biology-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Secondary Structure
Protein secondary structure is the three dimensional conformational isomerism, form of ''local segments'' of proteins. The two most common Protein structure#Secondary structure, secondary structural elements are alpha helix, alpha helices and beta sheets, though beta turns and omega loops occur as well. Secondary structure elements typically spontaneously form as an intermediate before the protein protein folding, folds into its three dimensional protein tertiary structure, tertiary structure. Secondary structure is formally defined by the pattern of hydrogen bonds between the Amine, amino hydrogen and carboxyl oxygen atoms in the peptide backbone chain, backbone. Secondary structure may alternatively be defined based on the regular pattern of backbone Dihedral angle#Dihedral angles of proteins, dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. The concept of secondary structure was first introduced by Kaj Ulrik ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequence Conservation
In evolutionary biology, conserved sequences are identical or similar sequences in nucleic acids ( DNA and RNA) or proteins across species ( orthologous sequences), or within a genome ( paralogous sequences), or between donor and receptor taxa ( xenologous sequences). Conservation indicates that a sequence has been maintained by natural selection. A highly conserved sequence is one that has remained relatively unchanged far back up the phylogenetic tree, and hence far back in geological time. Examples of highly conserved sequences include the RNA components of ribosomes present in all domains of life, the homeobox sequences widespread amongst Eukaryotes, and the tmRNA in Bacteria. The study of sequence conservation overlaps with the fields of genomics, proteomics, evolutionary biology, phylogenetics, bioinformatics and mathematics. History The discovery of the role of DNA in heredity, and observations by Frederick Sanger of variation between animal insulins in 1949, promp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cis-regulatory Element
''Cis''-regulatory elements (CREs) or ''Cis''-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology. CREs are found in the vicinity of the genes that they regulate. CREs typically regulate gene transcription by binding to transcription factors. A single transcription factor may bind to many CREs, and hence control the expression of many genes ( pleiotropy). The Latin prefix ''cis'' means "on this side", i.e. on the same molecule of DNA as the gene(s) to be transcribed. CRMs are stretches of DNA, usually 100–1000 DNA base pairs in length, where a number of transcription factors can bind and regulate expression of nearby genes and regulate their transcription rates. They are labeled as ''cis'' because they are ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bacteria
Bacteria (; singular: bacterium) are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria are vital in many stages of the nutrient cycle by recycling nutrients such as the fixation of nitrogen from the atmosphere. The nutrient cycle includes the decomposition of dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy. Bacteria also live in symbiotic and parasitic relationsh ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Upstream And Downstream (DNA)
In molecular biology and genetics, upstream and downstream both refer to relative positions of genetic code in DNA or RNA. Each strand of DNA or RNA has a 5' end and a 3' end, so named for the carbon position on the deoxyribose (or ribose) ring. By convention, upstream and downstream relate to the 5' to 3' direction respectively in which RNA transcription takes place. Upstream is toward the 5' end of the RNA molecule and downstream is toward the 3' end. When considering double-stranded DNA, upstream is toward the 5' end of the coding strand for the gene in question and downstream is toward the 3' end. Due to the anti-parallel nature of DNA, this means the 3' end of the template strand is upstream of the gene and the 5' end is downstream. Some genes on the same DNA molecule may be transcribed in opposite directions. This means the upstream and downstream areas of the molecule may change depending on which gene is used as the reference. See also *Upstream and downstream (transdu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
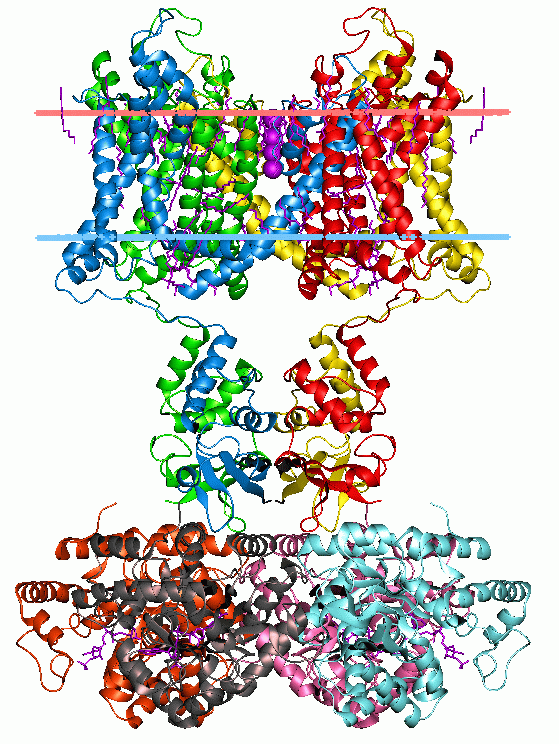
Membrane Protein
Membrane proteins are common proteins that are part of, or interact with, biological membranes. Membrane proteins fall into several broad categories depending on their location. Integral membrane proteins are a permanent part of a cell membrane and can either penetrate the membrane (transmembrane) or associate with one or the other side of a membrane ( integral monotopic). Peripheral membrane proteins are transiently associated with the cell membrane. Membrane proteins are common, and medically important—about a third of all human proteins are membrane proteins, and these are targets for more than half of all drugs. Nonetheless, compared to other classes of proteins, determining membrane protein structures remains a challenge in large part due to the difficulty in establishing experimental conditions that can preserve the correct conformation of the protein in isolation from its native environment. Function Membrane proteins perform a variety of functions vital to the sur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha-proteobacteria
Alphaproteobacteria is a class of bacteria in the phylum Pseudomonadota (formerly Proteobacteria). The Magnetococcales and Mariprofundales are considered basal or sister to the Alphaproteobacteria. The Alphaproteobacteria are highly diverse and possess few commonalities, but nevertheless share a common ancestor. Like all ''Proteobacteria'', its members are gram-negative and some of its intracellular parasitic members lack peptidoglycan and are consequently gram variable. Characteristics The Alphaproteobacteria are a diverse taxon and comprises several phototrophic genera, several genera metabolising C1-compounds (''e.g.'', ''Methylobacterium'' spp.), symbionts of plants (''e.g.'', ''Rhizobium'' spp.), endosymbionts of arthropods (''Wolbachia'') and intracellular pathogens (''e.g. Rickettsia''). Moreover, the class is sister to the protomitochondrion, the bacterium that was engulfed by the eukaryotic ancestor and gave rise to the mitochondria, which are organelles in eukaryotic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SpeF Leader
SpeF is a putative ''cis''-acting element identified in several gram negative alpha proteobacteria. It is proposed to be involved in regulating expression of genes involved in polyamide biosynthesis. SpeF is one of five putative regulatory elements identified by a computational screen of ''Agrobacterium tumefaciens'' and other alpha-proteobacterial genomes for conserved sequence motifs in operon leaders. In the majority of species analysed it is located in the leader of an operon containing the speF gene an ornithine decarboxylase enzyme that catalyses one of the first steps in polyamine biosynthesis. However, the authors did not detect binding of metabolites related to this pathway (L-ornithine, L-lysine, meso-diaminopimelate, putrescine, cadaverine, or spermidine Spermidine is a polyamine compound () found in ribosomes and living tissues and having various metabolic functions within organisms. It was originally isolated from semen. Function Spermidine is an aliphatic pol ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
SuhB
''suhB'', also known as ''mmgR'' (makes more granules regulator), is a non-coding RNA found multiple times in the ''Agrobacterium tumefaciens'' genome and related alpha-proteobacteria. Other non-coding RNAs uncovered in the same analysis include '' speF'', '' ybhL'', ''metA'', and ''serC''. Several studies in '' Sinorhizobium meliloti'' showed that the ''suhB'' element is indeed a non-coding RNA. It was first detected by Northern blot and called ''Sm8RNA'', then in an RNAseq study and referred to as ''SmelC689''. The mutant (lacking the small RNA) phenotype's cytoplasm contains a higher content of polyhydroxybutyrate (PBH) storage granules than the wild type strain. The sRNA is required to limit the PBH intracellular accumulation when the nitrogen-fixing '' Sinorhizobium meliloti'' is converting surplus carbon to nitrogen his needs to be modified, carbon cannot be converted to nitrogen/sup>. Further study confirmed that ''suhB'' fine-tunes the regulation of PBH storage. North ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |