|
Whole-exome Sequencing
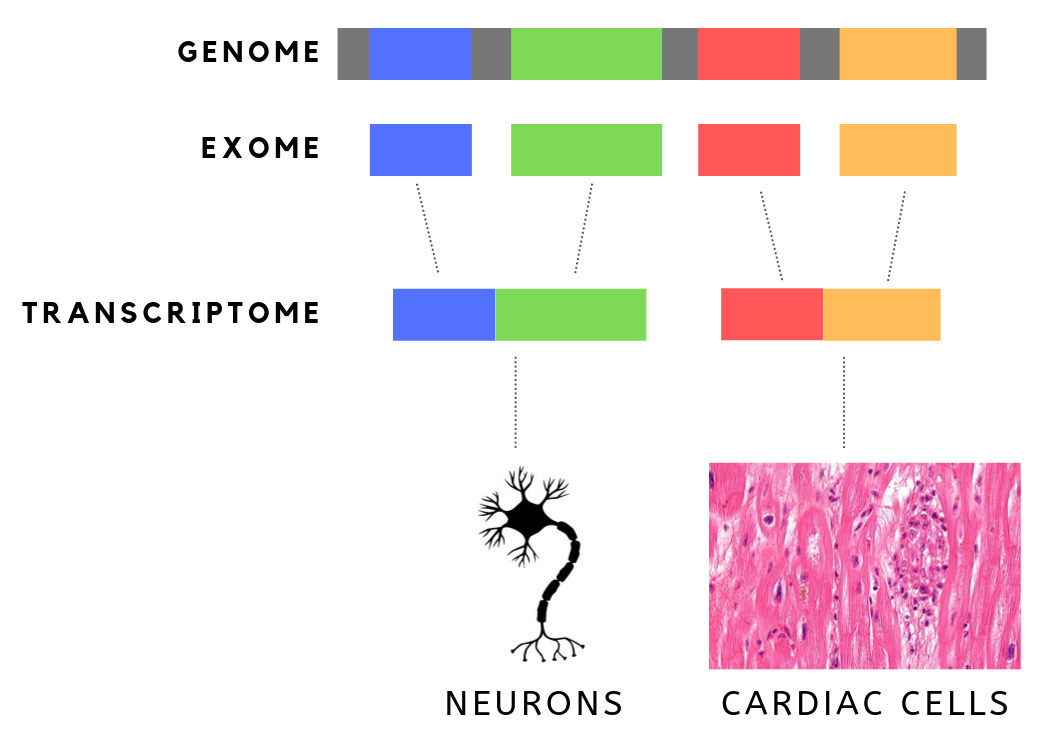
Exome sequencing, also known as whole exome sequencing (WES), is a genomic technique for sequencing all of the protein-coding regions of genes in a genome (known as the exome). It consists of two steps: the first step is to select only the subset of DNA that encodes proteins. These regions are known as exons—humans have about 180,000 exons, constituting about 1% of the human genome, or approximately 30 million base pairs. The second step is to sequence the exonic DNA using any high-throughput DNA sequencing technology. The goal of this approach is to identify genetic variants that alter protein sequences, and to do this at a much lower cost than whole-genome sequencing. Since these variants can be responsible for both Mendelian and common polygenic diseases, such as Alzheimer's disease, whole exome sequencing has been applied both in academic research and as a clinical diagnostic. Motivation and comparison to other approaches Exome sequencing is especially effective in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Exome Sequencing Workflow 1a
The exome is composed of all of the exons within the genome, the sequences which, when transcribed, remain within the mature RNA after introns are removed by RNA splicing. This includes untranslated regions of messenger RNA (mRNA), and coding regions. Exome sequencing has proven to be an efficient method of determining the genetic basis of more than two dozen Mendelian or single gene disorders. Statistics The human exome consists of roughly 233,785 exons, about 80% of which are less than 200 base pairs in length, constituting a total of about 1.1% of the total genome, or about 30 megabases of DNA. Though composing a very small fraction of the genome, mutations in the exome are thought to harbor 85% of mutations that have a large effect on disease. Definition It is important to note that the exome is distinct from the transcriptome, which is all of the transcribed RNA within a cell type. While the exome is constant from cell-type to cell-type, the transcriptome changes bas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Whole Genome Sequencing
Whole genome sequencing (WGS), also known as full genome sequencing, complete genome sequencing, or entire genome sequencing, is the process of determining the entirety, or nearly the entirety, of the DNA sequence of an organism's genome at a single time. This entails sequencing all of an organism's chromosomal DNA as well as DNA contained in the mitochondrial DNA, mitochondria and, for plants, in the chloroplast. Whole genome sequencing has largely been used as a research tool, but was being introduced to clinics in 2014. In the future of personalized medicine, whole genome sequence data may be an important tool to guide therapeutic intervention. The tool of DNA sequencing, gene sequencing at Single-nucleotide polymorphism, SNP level is also used to pinpoint functional variants from association studies and improve the knowledge available to researchers interested in evolutionary biology, and hence may lay the foundation for predicting disease susceptibility and drug response. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rare Variant (genetics)
A rare functional variant is a genetic variant which alters gene function, and which occurs at low frequency in a population. Rare variants play a significant role in both complex and Mendelian disease and are responsible for a portion of the missing heritability of complex diseases. The theoretical case for a significant role of rare variants is that alleles that strongly predispose an individual to disease will be kept at low frequencies in populations by purifying selection. Rare variants are increasingly being studied, as a consequence of whole exome and whole genome sequencing efforts. While these variants are individually infrequent in populations, there are many in human populations, and they can be unique to specific populations. They are more likely to be deleterious than common variants, as a result of rapid population growth and weak purifying selection. They have been suspected of acting independently or along with common variants to cause disease states. Methods ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genotyping
Genotyping is the process of determining differences in the genetic make-up ( genotype) of an individual by examining the individual's DNA sequence using biological assays and comparing it to another individual's sequence or a reference sequence. It reveals the alleles an individual has inherited from their parents. Traditionally genotyping is the use of DNA sequences to define biological populations by use of molecular tools. It does not usually involve defining the genes of an individual. Techniques Current methods of genotyping include restriction fragment length polymorphism identification (RFLPI) of genomic DNA, random amplified polymorphic detection (RAPD) of genomic DNA, amplified fragment length polymorphism detection (AFLPD), polymerase chain reaction (PCR), DNA sequencing, allele specific oligonucleotide (ASO) probes, and hybridization to DNA microarrays or beads. Genotyping is important in research of genes and gene variants associated with disease. Due to cur ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microarrays
A microarray is a multiplex lab-on-a-chip. Its purpose is to simultaneously detect the expression of thousands of genes from a sample (e.g. from a tissue). It is a two-dimensional array on a solid substrate—usually a glass slide or silicon thin-film cell—that assays (tests) large amounts of biological material using high-throughput screening miniaturized, multiplexed and parallel processing and detection methods. The concept and methodology of microarrays was first introduced and illustrated in antibody microarrays (also referred to as antibody matrix) by Tse Wen Chang in 1983 in a scientific publication and a series of patents. The "gene chip" industry started to grow significantly after the 1995 ''Science Magazine'' article by the Ron Davis and Pat Brown labs at Stanford University. With the establishment of companies, such as Affymetrix, Agilent, Applied Microarrays, Arrayjet, Illumina, and others, the technology of DNA microarrays has become the most sophisticated and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Whole-genome Sequencing
Whole genome sequencing (WGS), also known as full genome sequencing, complete genome sequencing, or entire genome sequencing, is the process of determining the entirety, or nearly the entirety, of the DNA sequence of an organism's genome at a single time. This entails sequencing all of an organism's chromosomal DNA as well as DNA contained in the mitochondria and, for plants, in the chloroplast. Whole genome sequencing has largely been used as a research tool, but was being introduced to clinics in 2014. In the future of personalized medicine, whole genome sequence data may be an important tool to guide therapeutic intervention. The tool of gene sequencing at SNP level is also used to pinpoint functional variants from association studies and improve the knowledge available to researchers interested in evolutionary biology, and hence may lay the foundation for predicting disease susceptibility and drug response. Whole genome sequencing should not be confused with DNA profilin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microarray
A microarray is a multiplex lab-on-a-chip. Its purpose is to simultaneously detect the expression of thousands of genes from a sample (e.g. from a tissue). It is a two-dimensional array on a solid substrate—usually a glass slide or silicon thin-film cell—that assays (tests) large amounts of biological material using high-throughput screening miniaturized, multiplexed and parallel processing and detection methods. The concept and methodology of microarrays was first introduced and illustrated in antibody microarrays (also referred to as antibody matrix) by Tse Wen Chang in 1983 in a scientific publication and a series of patents. The "gene chip" industry started to grow significantly after the 1995 ''Science Magazine'' article by the Ron Davis and Pat Brown labs at Stanford University. With the establishment of companies, such as Affymetrix, Agilent, Applied Microarrays, Arrayjet, Illumina, and others, the technology of DNA microarrays has become the most sophisticated and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Illumina (company)
Illumina, Inc. is an American biotechnology company, headquartered in San Diego, California. Incorporated on April 1, 1998, Illumina develops, manufactures, and markets integrated systems for the analysis of genetic variation and biological function. The company provides a line of products and services that serves the sequencing, genotyping and gene expression, and proteomics markets. Illumina's technology had purportedly reduced the cost of sequencing a human genome to by 2014. Its customers include genomic research centers, pharmaceutical companies, academic institutions, clinical research organizations, and biotechnology companies. History Illumina was founded in April 1998 by David Walt, Larry Bock, John Stuelpnagel, Anthony Czarnik, and Mark Chee. While working with CW Group, a venture-capital firm, Bock and Stuelpnagel uncovered what would become Illumina's BeadArray technology at Tufts University and negotiated an exclusive license to that technology. In 1999, Illumin ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Life Technologies (Thermo Fisher Scientific)
Life Technologies Corporation was a biotech company founded in November 2008 through a US $6.7billion merger of Invitrogen Corporation and Applied Biosystems Inc. The joint sales of the combined companies were about $3.5 billion; they had about 9,500 employees and owned more than 3,600 licenses and patents. Company name The name "Life Technologies" was an old name from the history of Invitrogen. GIBCO (Grand Island Biological Company) had been founded around 1960 in New York; in 1983 GIBCO merged with a reagent company called Bethesda Research Laboratories and the merged company was named Life Technologies. In 2000, Invitrogen acquired Life Technologies and discontinued that name. When Invitrogen and Applied Biosystems merged, the companies revived the name. The use of the "Life Technologies" brand name is disputed. Life Technologies (India) Private Limited, a company founded in 2002, operating in this corporate name claims ownership of the brand name. The "Life Technologies" ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Roche Applied Science
Roche Applied Science was a global business entity in the biotechnology sector that produces reagents and systems for life sciences research, with an emphasis on molecular genetics and cell biology research needs. Scope of business Roche Applied Science produced reagents and systems for related to DNA sequencing, microarrays, gene expression, and cell analysis. Important fields of application of the company's products are the research and development areas of the biotechnology and pharmaceuticals, pharmaceutical industries. The company also offers reagents and systems for the pharmaceuticals and diagnostics industries. Locations The company's headquarters was in Penzberg in Upper Bavaria, Germany; other main sites are Mannheim, Germany, Rotkreuz in Switzerland, Madison, WI, Pleasanton, CA, and Branford, CT in the United States. History Roche Applied Science was founded in 1859 as Boehringer Mannheim, a developer of rare chemicals and pharmaceuticals. In the 1950s, the bioreagen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oligonucleotides
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small bits of nucleic acids can be manufactured as single-stranded molecules with any user-specified sequence, and so are vital for artificial gene synthesis, polymerase chain reaction (PCR), DNA sequencing, molecular cloning and as molecular probes. In nature, oligonucleotides are usually found as small RNA molecules that function in the regulation of gene expression (e.g. microRNA), or are degradation intermediates derived from the breakdown of larger nucleic acid molecules. Oligonucleotides are characterized by the sequence of nucleotide residues that make up the entire molecule. The length of the oligonucleotide is usually denoted by " -mer" (from Greek ''meros'', "part"). For example, an oligonucleotide of six nucleotides (nt) is a hexamer, while one of 25 nt wou ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is a method widely used to rapidly make millions to billions of copies (complete or partial) of a specific DNA sample, allowing scientists to take a very small sample of DNA and amplify it (or a part of it) to a large enough amount to study in detail. PCR was invented in 1983 by the American biochemist Kary Mullis at Cetus Corporation; Mullis and biochemist Michael Smith (chemist), Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993. PCR is fundamental to many of the procedures used in genetic testing and research, including analysis of Ancient DNA, ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory research for a broad variety of applications ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
.jpg)
