|
Exome Sequencing Workflow 1a
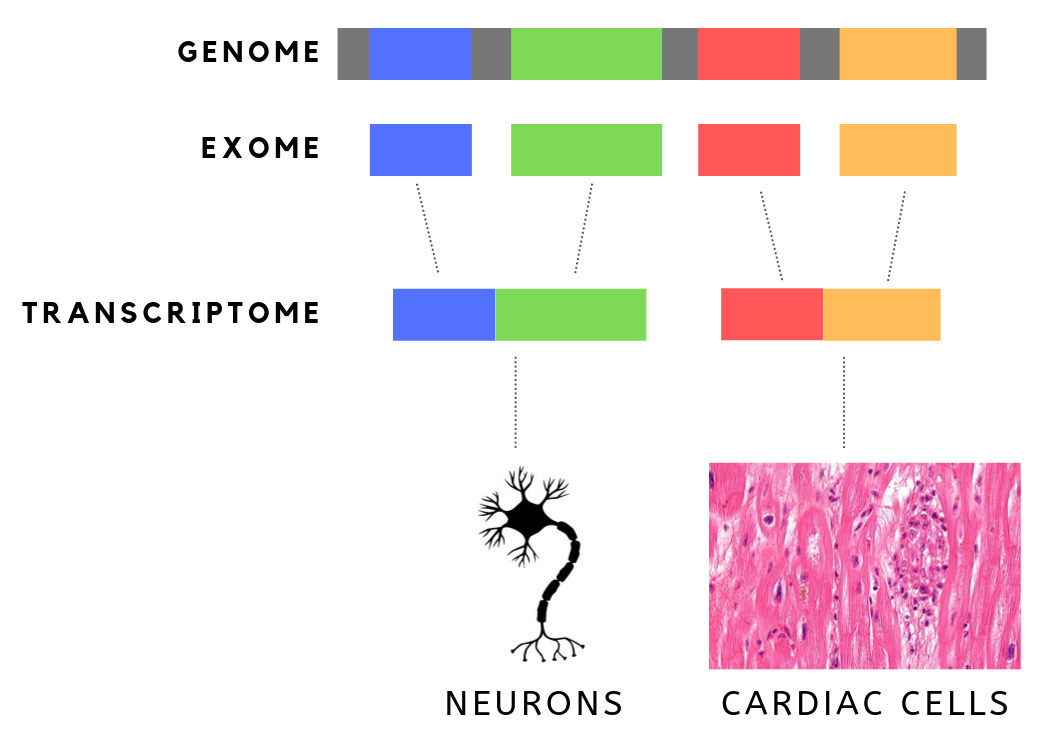
The exome is composed of all of the exons within the genome, the sequences which, when transcribed, remain within the mature RNA after introns are removed by RNA splicing. This includes untranslated regions of messenger RNA (mRNA), and coding regions. Exome sequencing has proven to be an efficient method of determining the genetic basis of more than two dozen Mendelian or single gene disorders. Statistics The human exome consists of roughly 233,785 exons, about 80% of which are less than 200 base pairs in length, constituting a total of about 1.1% of the total genome, or about 30 megabases of DNA. Though composing a very small fraction of the genome, mutations in the exome are thought to harbor 85% of mutations that have a large effect on disease. Definition It is important to note that the exome is distinct from the transcriptome, which is all of the transcribed RNA within a cell type. While the exome is constant from cell-type to cell-type, the transcriptome changes bas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Exon
An exon is any part of a gene that will form a part of the final mature RNA produced by that gene after introns have been removed by RNA splicing. The term ''exon'' refers to both the DNA sequence within a gene and to the corresponding sequence in RNA transcripts. In RNA splicing, introns are removed and exons are covalently joined to one another as part of generating the mature RNA. Just as the entire set of genes for a species constitutes the genome, the entire set of exons constitutes the exome. History The term ''exon'' derives from the expressed region and was coined by American biochemist Walter Gilbert in 1978: "The notion of the cistron… must be replaced by that of a transcription unit containing regions which will be lost from the mature messengerwhich I suggest we call introns (for intragenic regions)alternating with regions which will be expressedexons." This definition was originally made for protein-coding transcripts that are spliced before being translated. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Next-generation Sequencing
Massive parallel sequencing or massively parallel sequencing is any of several high-throughput approaches to DNA sequencing using the concept of massively parallel processing; it is also called next-generation sequencing (NGS) or second-generation sequencing. Some of these technologies emerged between 1994 and 1998 and have been commercially available since 2005. These technologies use miniaturized and parallelized platforms for sequencing of 1 million to 43 billion short reads (50 to 400 bases each) per instrument run. Many NGS platforms differ in engineering configurations and sequencing chemistry. They share the technical paradigm of massive parallel sequencing via spatially separated, clonally amplified DNA templates or single DNA molecules in a flow cytometry, flow cell. This design is very different from that of Sanger sequencing—also known as capillary sequencing or first-generation sequencing—which is based on electrophoretic separation of chain-termination products produ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mendelian Inheritance
Mendelian inheritance (also known as Mendelism) is a type of biological inheritance following the principles originally proposed by Gregor Mendel in 1865 and 1866, re-discovered in 1900 by Hugo de Vries and Carl Correns, and later popularized by William Bateson. These principles were initially controversial. When Mendel's theories were integrated with the Boveri–Sutton chromosome theory of inheritance by Thomas Hunt Morgan in 1915, they became the core of classical genetics. Ronald Fisher combined these ideas with the theory of natural selection in his 1930 book ''The Genetical Theory of Natural Selection'', putting evolution onto a mathematical footing and forming the basis for population genetics within the modern evolutionary synthesis. History The principles of Mendelian inheritance were named for and first derived by Gregor Johann Mendel, a nineteenth-century Moravian monk who formulated his ideas after conducting simple hybridisation experiments with pea plants (' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Mendelian Disease
A genetic disorder is a health problem caused by one or more abnormalities in the genome. It can be caused by a mutation in a single gene (monogenic) or multiple genes (polygenic) or by a chromosomal abnormality. Although polygenic disorders are the most common, the term is mostly used when discussing disorders with a single genetic cause, either in a gene or chromosome. The mutation responsible can occur spontaneously before embryonic development (a ''de novo'' mutation), or it can be inherited from two parents who are carriers of a faulty gene (autosomal recessive inheritance) or from a parent with the disorder (autosomal dominant inheritance). When the genetic disorder is inherited from one or both parents, it is also classified as a hereditary disease. Some disorders are caused by a mutation on the X chromosome and have X-linked inheritance. Very few disorders are inherited on the Y chromosome or mitochondrial DNA (due to their size). There are well over 6,000 known genet ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Structural Variation
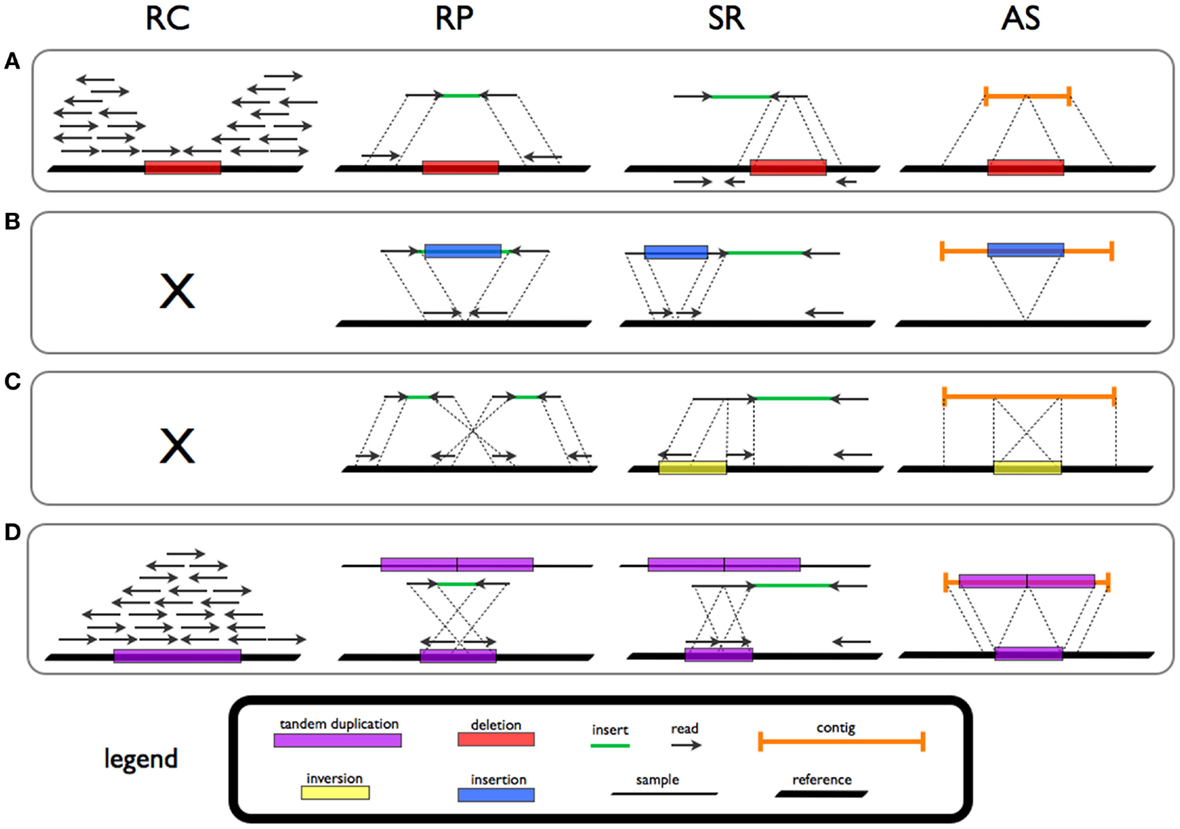
Genomic structural variation is the variation in structure of an organism's chromosome. It consists of many kinds of variation in the genome of one species, and usually includes microscopic and submicroscopic types, such as deletions, duplications, copy-number variants, insertions, inversions and translocations. Originally, a structure variation affects a sequence length about 1kb to 3Mb, which is larger than SNPs and smaller than chromosome abnormality (though the definitions have some overlap). However, the operational range of structural variants has widened to include events > 50bp. The definition of structural variation does not imply anything about frequency or phenotypical effects. Many structural variants are associated with genetic diseases, however many are not. Recent research about SVs indicates that SVs are more difficult to detect than SNPs. Approximately 13% of the human genome is defined as structurally variant in the normal population, and there are at least 2 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Sequencing
In genetics and biochemistry, sequencing means to determine the primary structure (sometimes incorrectly called the primary sequence) of an unbranched biopolymer. Sequencing results in a symbolic linear depiction known as a sequence which succinctly summarizes much of the atomic-level structure of the sequenced molecule. DNA sequencing DNA sequencing is the process of determining the nucleotide order of a given DNA fragment. So far, most DNA sequencing has been performed using the chain termination method developed by Frederick Sanger. This technique uses sequence-specific termination of a DNA synthesis reaction using modified nucleotide substrates. However, new sequencing technologies such as pyrosequencing are gaining an increasing share of the sequencing market. More genome data are now being produced by pyrosequencing than Sanger DNA sequencing. Pyrosequencing has enabled rapid genome sequencing. Bacterial genomes can be sequenced in a single run with several times cover ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Non-coding Region
Non-coding DNA (ncDNA) sequences are components of an organism's DNA that do not encode protein sequences. Some non-coding DNA is transcribed into functional non-coding RNA molecules (e.g. transfer RNA, microRNA, piRNA, ribosomal RNA, and regulatory RNAs). Other functional regions of the non-coding DNA fraction include regulatory sequences that control gene expression; scaffold attachment regions; origins of DNA replication; centromeres; and telomeres. Some non-coding regions appear to be mostly nonfunctional such as introns, pseudogenes, intergenic DNA, and fragments of transposons and viruses. Fraction of non-coding genomic DNA In bacteria, the coding regions typically take up 88 % of the genome. The remaining 12 % consists largely of non-coding genes and regulatory sequences, which means that almost all of the bacterial genome has a function. The amount of coding DNA in eukaryrotes is usually a much smaller fraction of the genome because eukaryotic genomes contain ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Single Nucleotide Variants
In genetics, a single-nucleotide polymorphism (SNP ; plural SNPs ) is a germline substitution of a single nucleotide at a specific position in the genome. Although certain definitions require the substitution to be present in a sufficiently large fraction of the population (e.g. 1% or more), many publications do not apply such a frequency threshold. For example, at a specific base position in the human genome, the G nucleotide may appear in most individuals, but in a minority of individuals, the position is occupied by an A. This means that there is a SNP at this specific position, and the two possible nucleotide variations – G or A – are said to be the alleles for this specific position. SNPs pinpoint differences in our susceptibility to a wide range of diseases, for example age-related macular degeneration (a common SNP in the CFH gene is associated with increased risk of the disease) or nonalcoholic fatty liver disease (a SNP in the PNPLA3 gene is associated with incre ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Whole Genome Sequencing
Whole genome sequencing (WGS), also known as full genome sequencing, complete genome sequencing, or entire genome sequencing, is the process of determining the entirety, or nearly the entirety, of the DNA sequence of an organism's genome at a single time. This entails sequencing all of an organism's chromosomal DNA as well as DNA contained in the mitochondrial DNA, mitochondria and, for plants, in the chloroplast. Whole genome sequencing has largely been used as a research tool, but was being introduced to clinics in 2014. In the future of personalized medicine, whole genome sequence data may be an important tool to guide therapeutic intervention. The tool of DNA sequencing, gene sequencing at Single-nucleotide polymorphism, SNP level is also used to pinpoint functional variants from association studies and improve the knowledge available to researchers interested in evolutionary biology, and hence may lay the foundation for predicting disease susceptibility and drug response. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microarrays
A microarray is a multiplex lab-on-a-chip. Its purpose is to simultaneously detect the expression of thousands of genes from a sample (e.g. from a tissue). It is a two-dimensional array on a solid substrate—usually a glass slide or silicon thin-film cell—that assays (tests) large amounts of biological material using high-throughput screening miniaturized, multiplexed and parallel processing and detection methods. The concept and methodology of microarrays was first introduced and illustrated in antibody microarrays (also referred to as antibody matrix) by Tse Wen Chang in 1983 in a scientific publication and a series of patents. The "gene chip" industry started to grow significantly after the 1995 ''Science Magazine'' article by the Ron Davis and Pat Brown labs at Stanford University. With the establishment of companies, such as Affymetrix, Agilent, Applied Microarrays, Arrayjet, Illumina, and others, the technology of DNA microarrays has become the most sophisticated and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Karyotyping
A karyotype is the general appearance of the complete set of metaphase chromosomes in the cells of a species or in an individual organism, mainly including their sizes, numbers, and shapes. Karyotyping is the process by which a karyotype is discerned by determining the chromosome complement of an individual, including the number of chromosomes and any abnormalities. A karyogram or idiogram is a graphical depiction of a karyotype, wherein chromosomes are organized in pairs, ordered by size and position of centromere for chromosomes of the same size. Karyotyping generally combines light microscopy and photography, and results in a photomicrographic (or simply micrographic) karyogram. In contrast, a schematic karyogram is a designed graphic representation of a karyotype. In schematic karyograms, just one of the sister chromatids of each chromosome is generally shown for brevity, and in reality they are generally so close together that they look as one on photomicrographs as well u ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Disorder
A genetic disorder is a health problem caused by one or more abnormalities in the genome. It can be caused by a mutation in a single gene (monogenic) or multiple genes (polygenic) or by a chromosomal abnormality. Although polygenic disorders are the most common, the term is mostly used when discussing disorders with a single genetic cause, either in a gene or chromosome. The mutation responsible can occur spontaneously before embryonic development (a ''de novo'' mutation), or it can be Heredity, inherited from two parents who are carriers of a faulty gene (autosomal recessive inheritance) or from a parent with the disorder (autosomal dominant inheritance). When the genetic disorder is inherited from one or both parents, it is also classified as a hereditary disease. Some disorders are caused by a mutation on the X chromosome and have X-linked inheritance. Very few disorders are inherited on the Y linkage, Y chromosome or Mitochondrial disease#Causes, mitochondrial DNA (due to t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |