|
TMCO6
Transmembrane and coiled-coil domain 6, TMCO6, is a protein that in humans is encoded by the ''TMCO6'' gene with aliases of PRO1580, HQ1580 or FLJ39769.1. Gene The human TMCO6 is found on chromosome 5 (position 5q31.3). The entire gene spans 5568 base pairs on the positive strand of chromosome 5 (140019113-140024689bp) but is alternately spliced into different variants. There are three known variants for TMCO6. Variant 1 is the longest and variant 2 and 3 are spliced into shorter mRNA strands. There is also a predicted variants X1-X7. Expression TMCO6 is expressed in liver tissue and is found during the fetal stage of development in humans. Homology Orthologs of the TMCO6 protein have been found among sequenced organisms with the exception of invertebrates, fungi, plants and bacteria. Tmco6 (transmembrane and coiled-coil domains 6) - Rat Genome Database. This includes close primates to some distant fish. Graphs show the evolutionary rate of TMCO6 with respect to differ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Aliases
A pseudonym (; ) or alias () is a fictitious name that a person or group assumes for a particular purpose, which differs from their original or true name (orthonym). This also differs from a new name that entirely or legally replaces an individual's own. Many pseudonym holders use pseudonyms because they wish to remain anonymous, but anonymity is difficult to achieve and often fraught with legal issues. Scope Pseudonyms include stage names, user names, ring names, pen names, aliases, superhero or villain identities and code names, gamer identifications, and regnal names of emperors, popes, and other monarchs. In some cases, it may also include nicknames. Historically, they have sometimes taken the form of anagrams, Graecisms, and Latinisations. Pseudonyms should not be confused with new names that replace old ones and become the individual's full-time name. Pseudonyms are "part-time" names, used only in certain contexts – to provide a more clear-cut separation between one's p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chromosome 5
Chromosome 5 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 5 spans about 181 million base pairs (the building blocks of DNA) and represents almost 6% of the total DNA in cells. Chromosome 5 is the 5th largest human chromosome, yet has one of the lowest gene densities. This is partially explained by numerous gene-poor regions that display a remarkable degree of non-coding and syntenic conservation with non-mammalian vertebrates, suggesting they are functionally constrained. Because chromosome 5 is responsible for many forms of growth and development (cell divisions) changes may cause cancers. One example would be acute myeloid leukemia (AML). Genes Number of genes The following are some of the gene count estimates of human chromosome 5. Because researchers use different approaches to genome annotation their predictions of the number of genes on each chromosome varies (for technical details, see gene prediction ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
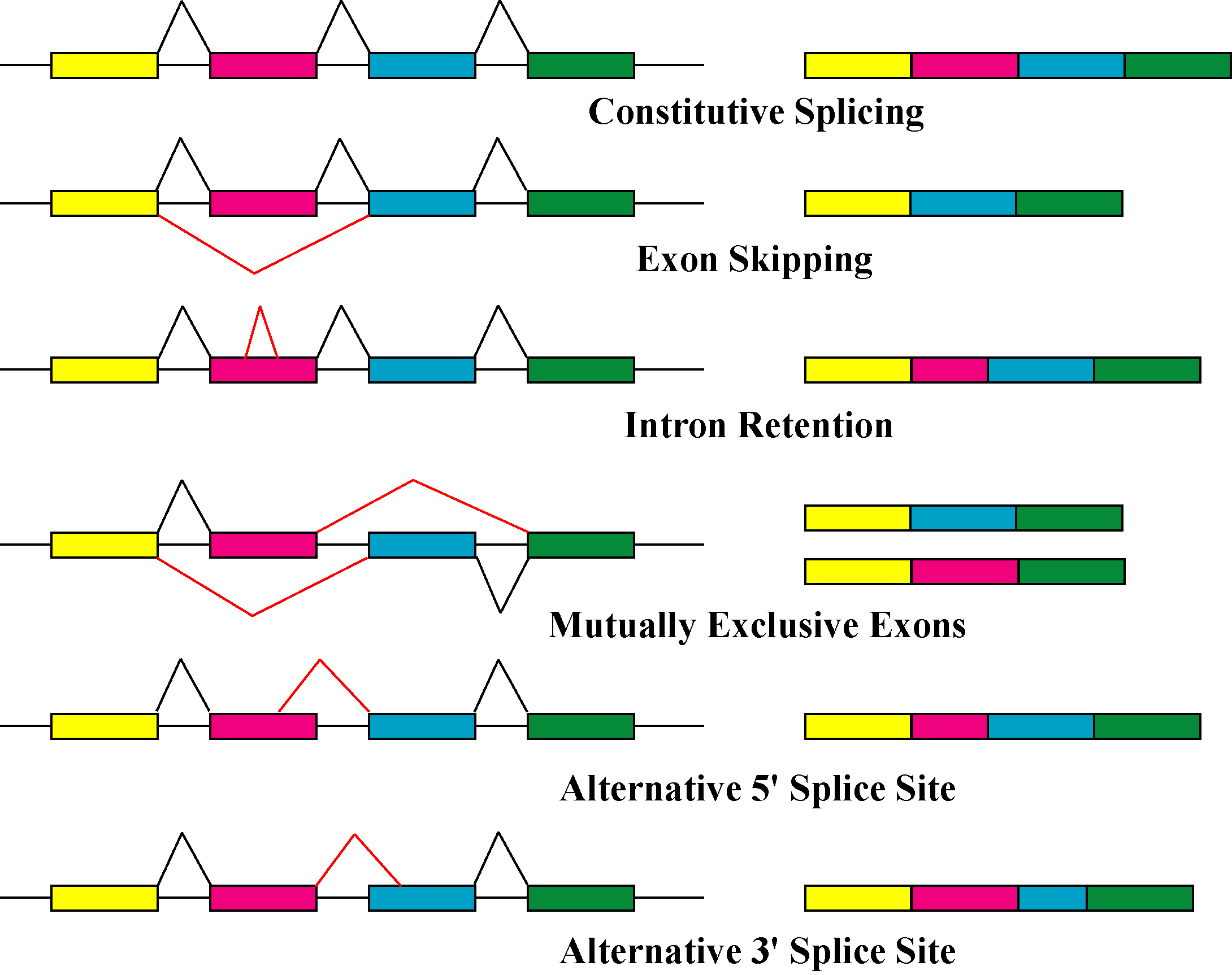
Isoform
A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene or gene family and are the result of genetic differences. While many perform the same or similar biological roles, some isoforms have unique functions. A set of protein isoforms may be formed from alternative splicings, variable promoter usage, or other post-transcriptional modifications of a single gene; post-translational modifications are generally not considered. (For that, see Proteoforms.) Through RNA splicing mechanisms, mRNA has the ability to select different protein-coding segments ( exons) of a gene, or even different parts of exons from RNA to form different mRNA sequences. Each unique sequence produces a specific form of a protein. The discovery of isoforms could explain the discrepancy between the small number of protein coding regions genes revealed by the human genome project and the large diversity of proteins seen in an organism: different ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Orthologs
Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a speciation event (orthologs), or a duplication event (paralogs), or else a horizontal (or lateral) gene transfer event (xenologs). Homology among DNA, RNA, or proteins is typically inferred from their nucleotide or amino acid sequence similarity. Significant similarity is strong evidence that two sequences are related by evolutionary changes from a common ancestral sequence. Alignments of multiple sequences are used to indicate which regions of each sequence are homologous. Identity, similarity, and conservation The term "percent homology" is often used to mean "sequence similarity”, that is the percentage of identical residues (''percent identity''), or the percentage of residues conserved with similar physicochemical properties ('' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fibrinogen
Fibrinogen (factor I) is a glycoprotein complex, produced in the liver, that circulates in the blood of all vertebrates. During tissue and vascular injury, it is converted enzymatically by thrombin to fibrin and then to a fibrin-based blood clot. Fibrin clots function primarily to occlude blood vessels to stop bleeding. Fibrin also binds and reduces the activity of thrombin. This activity, sometimes referred to as antithrombin I, limits clotting. Fibrin also mediates blood platelet and endothelial cell spreading, tissue fibroblast proliferation, capillary tube formation, and angiogenesis and thereby promotes revascularization and wound healing. Reduced and/or dysfunctional fibrinogens occur in various congenital and acquired human fibrinogen-related disorders. These disorders represent a group of rare conditions in which individuals may present with severe episodes of pathological bleeding and thrombosis; these conditions are treated by supplementing blood fibrinogen levels an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Evolutionary History
The history of life on Earth traces the processes by which living and fossil organisms evolved, from the earliest emergence of life to present day. Earth formed about 4.5 billion years ago (abbreviated as ''Ga'', for ''gigaannum'') and evidence suggests that life emerged prior to 3.7 Ga. Although there is some evidence of life as early as 4.1 to 4.28 Ga, it remains controversial due to the possible non-biological formation of the purported fossils. The similarities among all known present-day species indicate that they have diverged through the process of evolution from a common ancestor. Only a very small percentage of species have been identified: one estimate claims that Earth may have 1 trillion species. * However, only 1.75–1.8 million have been named "A version of this article appears in print on Nov. 9, 2014, Section SR, Page 6 of the New York edition with the headline: Prehistory's Brilliant Future." and 1.8 million documented in a central database. These currently ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Evolutionary Rate
The rate of evolution is quantified as the speed of genetic or morphological change in a lineage over a period of time. The speed at which a molecular entity (such as a protein, gene, etc.) evolves is of considerable interest in evolutionary biology since determining the evolutionary rate is the first step in characterizing its evolution. Calculating rates of evolutionary change is also useful when studying phenotypic changes in phylogenetic comparative biology. In either case, it can be beneficial to consider and compare both genomic (such as DNA sequence) data and paleontological (such as fossil record) data, especially in regards to estimating the timing of divergence events and establishing geological time scales. At the organism level In his extensive study of evolution and paleontology, George Gaylord Simpson established evolutionary rates by using the fossil record to count the number of successive genera that occurred within a lineage during a given time period. For examp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Isoforms
A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene or gene family and are the result of genetic differences. While many perform the same or similar biological roles, some isoforms have unique functions. A set of protein isoforms may be formed from alternative splicings, variable promoter usage, or other post-transcriptional modifications of a single gene; post-translational modifications are generally not considered. (For that, see Proteoforms.) Through RNA splicing mechanisms, mRNA has the ability to select different protein-coding segments ( exons) of a gene, or even different parts of exons from RNA to form different mRNA sequences. Each unique sequence produces a specific form of a protein. The discovery of isoforms could explain the discrepancy between the small number of protein coding regions genes revealed by the human genome project and the large diversity of proteins seen in an organism: different ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclear Localization Sequence
A nuclear localization signal ''or'' sequence (NLS) is an amino acid sequence that 'tags' a protein for import into the cell nucleus by nuclear transport. Typically, this signal consists of one or more short sequences of positively charged lysines or arginines exposed on the protein surface. Different nuclear localized proteins may share the same NLS. An NLS has the opposite function of a nuclear export signal (NES), which targets proteins out of the nucleus. Types Classical These types of NLSs can be further classified as either monopartite or bipartite. The major structural differences between the two are that the two basic amino acid clusters in bipartite NLSs are separated by a relatively short spacer sequence (hence bipartite - 2 parts), while monopartite NLSs are not. The first NLS to be discovered was the sequence PKKKRKV in the SV40 Large T-antigen (a monopartite NLS). The NLS of nucleoplasmin, KR AATKKAGQAKKK, is the prototype of the ubiquitous bipartite signal: two cluster ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Armadillo Repeat
An armadillo repeat is the name of a characteristic, repetitive amino acid sequence of about 40 residues in length that is found in many proteins. Proteins that contain armadillo repeats typically contain several tandemly repeated copies. Each armadillo repeat is composed of a pair of alpha helices that form a hairpin structure. Multiple copies of the repeat form what is known as an alpha solenoid structure. Examples of proteins that contain armadillo repeats include β-catenin, α-importin, plakoglobin, adenomatous polyposis coli (APC), and many others. The term armadillo derives from the historical name of the β-catenin gene in the fruitfly ''Drosophila'' where the armadillo repeat was first discovered. Although β-catenin was previously believed to be a protein involved in linking cadherin cell adhesion proteins to the cytoskeleton, recent work indicates that β-catenin regulates the homodimerization of alpha-catenin, which in turn controls actin branching and bundling.Nus ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Karyopherin Alpha 1
Importin subunit alpha-5 is a protein that in humans is encoded by the ''KPNA1'' gene. Interactions Importin subunit alpha-5 has been shown to interact with KPNB1 and UBR5 E3 ubiquitin-protein ligase UBR5 is an enzyme that in humans is encoded by the ''UBR5'' gene. Function This gene encodes a progestin-induced protein, which belongs to the HECT (homology to E6-AP carboxyl terminus) family. The HECT family protei .... References Further reading * * * * * * * * * * * * * * * * * * * Armadillo-repeat-containing proteins {{gene-3-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |