Isoform on:
[Wikipedia]
[Google]
[Amazon]
 A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single
A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single
isoform.io
The specificity of translated isoforms is derived by the protein's structure/function, as well as the cell type and developmental stage during which they are produced. Determining specificity becomes more complicated when a protein has multiple subunits and each subunit has multiple isoforms. For example, the 5' AMP-activated protein kinase (AMPK), an enzyme, which performs different roles in human cells, has 3 subunits: * α, catalytic domain, has two isoforms: α1 and α2 which are encoded from PRKAA1 and
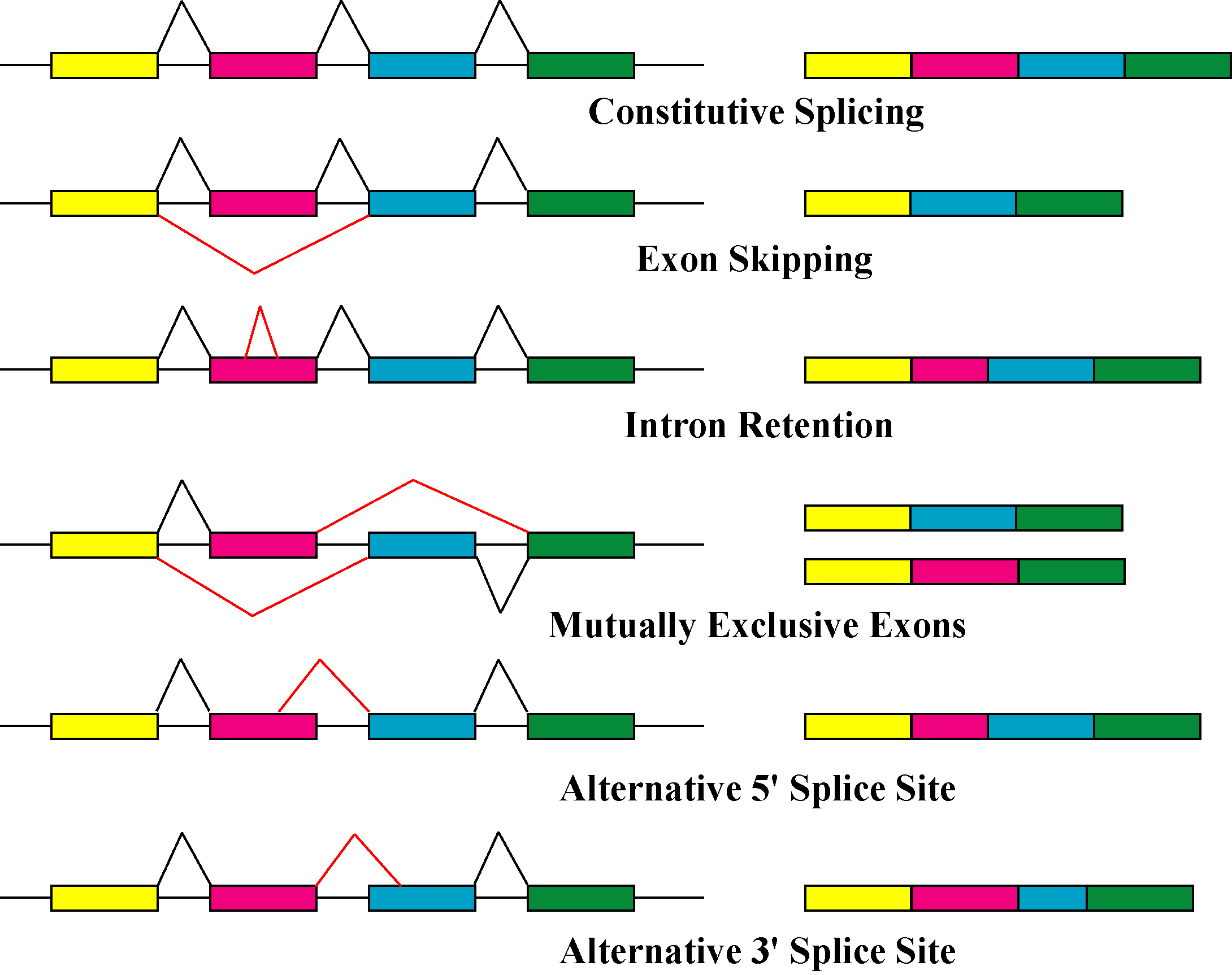 The primary mechanisms that produce protein isoforms are alternative splicing and variable promoter usage, though modifications due to genetic changes, such as mutations and polymorphisms are sometimes also considered distinct isoforms.
Alternative splicing is the main post-transcriptional modification process that produces mRNA transcript isoforms, and is a major molecular mechanism that may contribute to protein diversity. The spliceosome, a large
The primary mechanisms that produce protein isoforms are alternative splicing and variable promoter usage, though modifications due to genetic changes, such as mutations and polymorphisms are sometimes also considered distinct isoforms.
Alternative splicing is the main post-transcriptional modification process that produces mRNA transcript isoforms, and is a major molecular mechanism that may contribute to protein diversity. The spliceosome, a large
MeSH entry protein isoforms
Definitions Isoform
{{DEFAULTSORT:Protein Isoform Protein structure
 A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single
A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene
In biology, the word gene (from , ; "... Wilhelm Johannsen coined the word gene to describe the Mendelian units of heredity..." meaning ''generation'' or ''birth'' or ''gender'') can have several different meanings. The Mendelian gene is a b ...
or gene family and are the result of genetic differences. While many perform the same or similar biological roles, some isoforms have unique functions. A set of protein isoforms may be formed from alternative splicings, variable promoter usage, or other post-transcriptional modifications of a single gene; post-translational modification
Post-translational modification (PTM) is the covalent and generally enzymatic modification of proteins following protein biosynthesis. This process occurs in the endoplasmic reticulum and the golgi apparatus. Proteins are synthesized by ribos ...
s are generally not considered. (For that, see Proteoforms.) Through RNA splicing mechanisms, mRNA
In molecular biology, messenger ribonucleic acid (mRNA) is a single-stranded molecule of RNA that corresponds to the genetic sequence of a gene, and is read by a ribosome in the process of synthesizing a protein.
mRNA is created during the ...
has the ability to select different protein-coding segments ( exons) of a gene, or even different parts of exons from RNA to form different mRNA sequences. Each unique sequence produces a specific form of a protein.
The discovery of isoforms could explain the discrepancy between the small number of protein coding regions genes revealed by the human genome project and the large diversity of proteins seen in an organism: different proteins encoded by the same gene could increase the diversity of the proteome
The proteome is the entire set of proteins that is, or can be, expressed by a genome, cell, tissue, or organism at a certain time. It is the set of expressed proteins in a given type of cell or organism, at a given time, under defined conditions. ...
. Isoforms at the RNA level are readily characterized by cDNA transcript studies. Many human genes possess confirmed alternative splicing isoforms. It has been estimated that ~100,000 expressed sequence tags ( ESTs) can be identified in humans. Isoforms at the protein level can manifest in the deletion of whole domains or shorter loops, usually located on the surface of the protein.
Definition
One single gene has the ability to produce multiple proteins that differ both in structure and composition; this process is regulated by the alternative splicing of mRNA, though it is not clear to what extent such a process affects the diversity of the human proteome, as the abundance of mRNA transcript isoforms does not necessarily correlate with the abundance of protein isoforms. Three-dimensional protein structure comparisons can be used to help determine which, if any, isoforms represent functional protein products, and the structure of most isoforms in the human proteome has been predicted byAlphaFold
AlphaFold is an artificial intelligence (AI) program developed by DeepMind, a subsidiary of Alphabet, which performs predictions of protein structure. The program is designed as a deep learning system.
AlphaFold AI software has had two major ve ...
and publicly released aisoform.io
The specificity of translated isoforms is derived by the protein's structure/function, as well as the cell type and developmental stage during which they are produced. Determining specificity becomes more complicated when a protein has multiple subunits and each subunit has multiple isoforms. For example, the 5' AMP-activated protein kinase (AMPK), an enzyme, which performs different roles in human cells, has 3 subunits: * α, catalytic domain, has two isoforms: α1 and α2 which are encoded from PRKAA1 and
PRKAA2
5'-AMP-activated protein kinase catalytic subunit alpha-2 is an enzyme that in humans is encoded by the ''PRKAA2'' gene.
Function
The protein encoded by this gene is a catalytic subunit of the AMP-activated protein kinase (AMPK). AMPK is a h ...
* β, regulatory domain, has two isoforms: β1 and β2 which are encoded from PRKAB1 and PRKAB2
* γ, regulatory domain, has three isoforms: γ1, γ2, and γ3 which are encoded from PRKAG1, PRKAG2, and PRKAG3
In human skeletal muscle, the preferred form is α2β2γ1. But in the human liver, the most abundant form is α1β2γ1.
Mechanism
 The primary mechanisms that produce protein isoforms are alternative splicing and variable promoter usage, though modifications due to genetic changes, such as mutations and polymorphisms are sometimes also considered distinct isoforms.
Alternative splicing is the main post-transcriptional modification process that produces mRNA transcript isoforms, and is a major molecular mechanism that may contribute to protein diversity. The spliceosome, a large
The primary mechanisms that produce protein isoforms are alternative splicing and variable promoter usage, though modifications due to genetic changes, such as mutations and polymorphisms are sometimes also considered distinct isoforms.
Alternative splicing is the main post-transcriptional modification process that produces mRNA transcript isoforms, and is a major molecular mechanism that may contribute to protein diversity. The spliceosome, a large ribonucleoprotein
Nucleoproteins are proteins conjugated with nucleic acids (either DNA or RNA). Typical nucleoproteins include ribosomes, nucleosomes and viral nucleocapsid proteins.
Structures
Nucleoproteins tend to be positively charged, facilitating int ...
, is the molecular machine inside the nucleus responsible for RNA cleavage and ligation, removing non-protein coding segments (introns
An intron is any nucleotide sequence within a gene that is not expressed or operative in the final RNA product. The word ''intron'' is derived from the term ''intragenic region'', i.e. a region inside a gene."The notion of the cistron .e., gene ...
).
Because splicing is a process that occurs between transcription and translation
Translation is the communication of the meaning of a source-language text by means of an equivalent target-language text. The English language draws a terminological distinction (which does not exist in every language) between ''transla ...
, its primary effects have mainly been studied through genomics techniques—for example, microarray analyses and RNA sequencing have been used to identify alternatively spliced transcripts and measure their abundances. Transcript abundance is often used as a proxy for the abundance of protein isoforms, though proteomics experiments using gel electrophoresis and mass spectrometry have demonstrated that the correlation between transcript and protein counts is often low, and that one protein isoform is usually dominant. One 2015 study states that the cause of this discrepancy likely occurs after translation, though the mechanism is essentially unknown. Consequently, although alternative splicing has been implicated as an important link between variation and disease, there is no conclusive evidence that it acts primarily by producing novel protein isoforms.
Alternative splicing generally describes a tightly regulated process in which alternative transcripts are intentionally generated by the splicing machinery. However, such transcripts are also produced by splicing errors in a process called "noisy splicing," and are also potentially translated into protein isoforms. Although ~95% of multi-exonic genes are thought to be alternatively spliced, one study on noisy splicing observed that most of the different low-abundance transcripts are noise, and predicts that most alternative transcript and protein isoforms present in a cell are not functionally relevant.
Other transcriptional and post-transcriptional regulatory steps can also produce different protein isoforms. Variable promoter usage occurs when the transcriptional machinery of a cell ( RNA polymerase, transcription factors
In molecular biology, a transcription factor (TF) (or sequence-specific DNA-binding factor) is a protein that controls the rate of transcription of genetic information from DNA to messenger RNA, by binding to a specific DNA sequence. The fun ...
, and other enzymes) begin transcription at different promoters—the region of DNA near a gene that serves as an initial binding site—resulting in slightly modified transcripts and protein isoforms.
Characteristics
Generally, one protein isoform is labeled as the canonical sequence based on criteria such as its prevalence and similarity to orthologous—or functionally analogous—sequences in other species. Isoforms are assumed to have similar functional properties, as most have similar sequences, and share some to most exons with the canonical sequence. However, some isoforms show much greater divergence (for example, through trans-splicing), and can share few to no exons with the canonical sequence. In addition, they can have different biological effects—for example, in an extreme case, the function of one isoform can promote cell survival, while another promotes cell death—or can have similar basic functions but differ in their sub-cellular localization. A 2016 study, however, functionally characterized all the isoforms of 1,492 genes and determined that most isoforms behave as "functional alloforms." The authors came to the conclusion that isoforms behave like distinct proteins after observing that the functional of most isoforms did not overlap. Because the study was conducted on cells ''in vitro'', it is not known if the isoforms in the expressed human proteome share these characteristics. Additionally, because the function of each isoform must generally be determined separately, most identified and predicted isoforms still have unknown functions.Related Concept
Glycoform
A glycoform is an isoform of a protein that differs only with respect to the number or type of attached glycan.Glycoproteins
Glycoproteins are proteins which contain oligosaccharide chains covalently attached to amino acid side-chains. The carbohydrate is attached to the protein in a cotranslational or posttranslational modification. This process is known as glycosy ...
often consist of a number of different glycoforms, with alterations in the attached saccharide
In organic chemistry, a carbohydrate () is a biomolecule consisting of carbon (C), hydrogen (H) and oxygen (O) atoms, usually with a hydrogen–oxygen atom ratio of 2:1 (as in water) and thus with the empirical formula (where ''m'' may or ma ...
or oligosaccharide
An oligosaccharide (/ˌɑlɪgoʊˈsækəˌɹaɪd/; from the Greek ὀλίγος ''olígos'', "a few", and σάκχαρ ''sácchar'', "sugar") is a saccharide polymer containing a small number (typically two to ten) of monosaccharides (simple sug ...
. These modifications may result from differences in biosynthesis during the process of glycosylation, or due to the action of glycosidases
Glycoside hydrolases (also called glycosidases or glycosyl hydrolases) catalyze
Catalysis () is the process of increasing the rate of a chemical reaction by adding a substance known as a catalyst (). Catalysts are not consumed in the rea ...
or glycosyltransferases. Glycoforms may be detected through detailed chemical analysis of separated glycoforms, but more conveniently detected through differential reaction with lectins
Lectins are carbohydrate-binding proteins that are highly specific for sugar groups that are part of other molecules, so cause agglutination of particular cells or precipitation of glycoconjugates and polysaccharides. Lectins have a role in rec ...
, as in lectin affinity chromatography
Affinity chromatography is a method of separating a biomolecule from a mixture, based on a highly specific macromolecular binding interaction between the biomolecule and another substance. The specific type of binding interaction depends on the ...
and lectin affinity electrophoresis
Affinity electrophoresis is a general name for many analytical methods used in biochemistry and biotechnology. Both qualitative and quantitative information may be obtained through affinity electrophoresis. The methods include the so-called elect ...
. Typical examples of glycoproteins consisting of glycoforms are the blood proteins as orosomucoid
Orosomucoid (ORM) or alpha-1-acid glycoprotein (''α1AGp'', ''AGP'' or ''AAG'') is an acute phase protein found in plasma. It is an alpha-globulin glycoprotein and is modulated by two polymorphic genes. It is synthesized primarily in hepatoc ...
, antitrypsin, and haptoglobin
Haptoglobin (abbreviated as Hp) is the protein that in humans is encoded by the ''HP'' gene. In blood plasma, haptoglobin binds with high affinity to ''free'' hemoglobin released from erythrocytes, and thereby inhibits its deleterious oxidative ...
. An unusual glycoform variation is seen in neuronal cell adhesion molecule, NCAM involving polysialic acids, PSA.
Examples
*G-actin
Actin is a family of globular multi-functional proteins that form microfilaments in the cytoskeleton, and the thin filaments in muscle fibrils. It is found in essentially all eukaryotic cells, where it may be present at a concentration of over ...
: despite its conserved nature, it has a varying number of isoforms (at least six in mammals).
* Creatine kinase
Creatine kinase (CK), also known as creatine phosphokinase (CPK) or phosphocreatine kinase, is an enzyme () expressed by various tissues and cell types. CK catalyses the conversion of creatine and uses adenosine triphosphate (ATP) to create pho ...
, the presence of which in the blood can be used as an aid in the diagnosis of myocardial infarction
A myocardial infarction (MI), commonly known as a heart attack, occurs when blood flow decreases or stops to the coronary artery of the heart, causing damage to the heart muscle. The most common symptom is chest pain or discomfort which may ...
, exists in 3 isoforms.
* Hyaluronan synthase, the enzyme responsible for the production of hyaluronan, has three isoforms in mammalian cells.
* UDP-glucuronosyltransferase, an enzyme superfamily responsible for the detoxification pathway of many drugs, environmental pollutants, and toxic endogenous compounds has 16 known isoforms encoded in the human genome.
*G6PDA: normal ratio of active isoforms in cells of any tissue is 1:1 shared with G6PDG. This is precisely the normal isoform ratio in hyperplasia. Only one of these isoforms is found during neoplasia.Pathoma, Fundamentals of Pathology
Monoamine oxidase, a family of enzymes that catalyze the oxidation of monoamines, exists in two isoforms, MAO-A and MAO-B.
See also
* Gene isoformReferences
External links
MeSH entry protein isoforms
Definitions Isoform
{{DEFAULTSORT:Protein Isoform Protein structure