|
T-Coffee
T-Coffee (Tree-based Consistency Objective Function for Alignment Evaluation) is a multiple sequence alignment software using a progressive approach. It generates a library of pairwise alignments to guide the multiple sequence alignment. It can also combine multiple sequences alignments obtained previously and in the latest versions can use structural information from PDB files (3D-Coffee). It has advanced features to evaluate the quality of the alignments and some capacity for identifying occurrence of motifs (Mocca). It produces alignment in the aln format ( Clustal) by default, but can also produce PIR, MSF, and FASTA format. The most common input formats are supported ( FASTA, PIR). Algorithm T-Coffee algorithm consist of two main features, the first by utilizing heterogeneous data sources it is able to provide simple and flexible means of generating multiple alignments. T-coffee can compute multiple alignments using a library that was generated using a mixture of local and ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Multiple Sequence Alignment
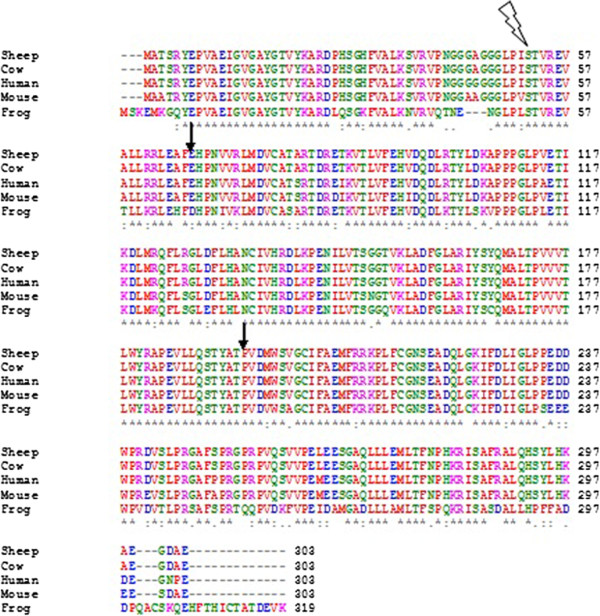
Multiple sequence alignment (MSA) may refer to the process or the result of sequence alignment of three or more biological sequences, generally protein, DNA, or RNA. In many cases, the input set of query sequences are assumed to have an evolutionary relationship by which they share a linkage and are descended from a common ancestor. From the resulting MSA, sequence homology can be inferred and phylogenetic analysis can be conducted to assess the sequences' shared evolutionary origins. Visual depictions of the alignment as in the image at right illustrate mutation events such as point mutations (single amino acid or nucleotide changes) that appear as differing characters in a single alignment column, and insertion or deletion mutations (indels or gaps) that appear as hyphens in one or more of the sequences in the alignment. Multiple sequence alignment is often used to assess sequence conservation of protein domains, tertiary and secondary structures, and even individual amino acid ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ClustalW
Clustal is a series of widely used computer programs used in bioinformatics for multiple sequence alignment. There have been many versions of Clustal over the development of the algorithm that are listed below. The analysis of each tool and its algorithm are also detailed in their respective categories. Available operating systems listed in the sidebar are a combination of the software availability and may not be supported for every current version of the Clustal tools. Clustal Omega has the widest variety of operating systems out of all the Clustal tools. History There have been many variations of the Clustal software, all of which are listed below: * Clustal: The original software for multiple sequence alignments, created by Des Higgins in 1988, was based on deriving phylogenetic trees from pairwise sequences of amino acids or nucleotides. * ClustalV: The second generation of the Clustal software was released in 1992 and was a rewrite of the original Clustal package. It in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Clustal
Clustal is a series of widely used computer programs used in bioinformatics for multiple sequence alignment. There have been many versions of Clustal over the development of the algorithm that are listed below. The analysis of each tool and its algorithm are also detailed in their respective categories. Available operating systems listed in the sidebar are a combination of the software availability and may not be supported for every current version of the Clustal tools. Clustal Omega has the widest variety of operating systems out of all the Clustal tools. History There have been many variations of the Clustal software, all of which are listed below: * Clustal: The original software for multiple sequence alignments, created by Des Higgins in 1988, was based on deriving phylogenetic trees from pairwise sequences of amino acids or nucleotides. * ClustalV: The second generation of the Clustal software was released in 1992 and was a rewrite of the original Clustal package. It int ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ProbCons
ProbCons is an open source probabilistic consistency-based multiple alignment of amino acid sequences. It is one of the most efficient protein multiple sequence alignment programs, since it has repeatedly demonstrated a statistically significant advantage in accuracy over similar tools, including Clustal and MAFFT. Algorithm The following describes the basic outline of the ProbCons algorithm. Step 1: Reliability of an alignment edge For every pair of sequences compute the probability that letters x_i and y_i are paired in a^* an alignment that is generated by the model. \begin P(x_i \sim y_i, x,y) & \stackrel Pr x,y\\ & = \sum_ Pr x,y\ & = \sum_ \mathbf\ Pr x,y\end (Where \mathbf\ is equal to 1 if x_i and y_i are in the alignment and 0 otherwise.) Step 2: Maximum expected accuracy The accuracy of an alignment a^* with respect to another alignment a is defined as the number of common aligned pairs divided by the length of the shorter sequence. Calculate expected accuracy of each ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
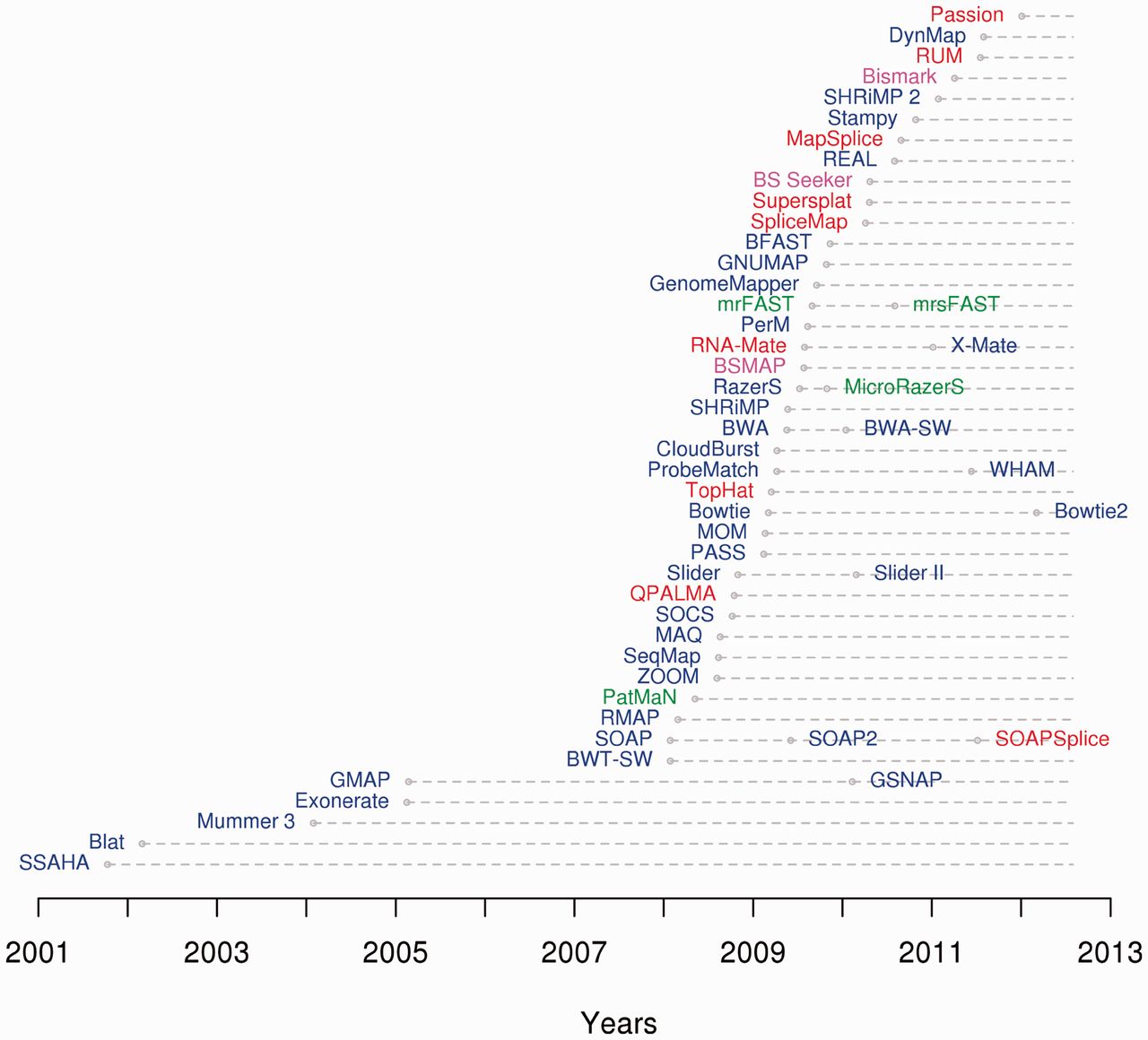
List Of Sequence Alignment Software
This list of sequence alignment software is a compilation of software tools and web portals used in pairwise sequence alignment and multiple sequence alignment. See structural alignment software for structural alignment of proteins. Database search only *Sequence type: protein or nucleotide Pairwise alignment *Sequence type: protein or nucleotide **Alignment type: local or global Multiple sequence alignment *Sequence type: protein or nucleotide. **Alignment type: local or global Genomics analysis *Sequence type: protein or nucleotide Motif finding *Sequence type: protein or nucleotide Benchmarking Alignment viewers, editors Please see List of alignment visualization software. Short-read sequence alignment See also * List of open source bioinformatics software References {{Reflist Genetics-related lists, Sequence Lists of bioinformatics software, Sequence alignment software ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FASTA
FASTA is a DNA and protein sequence alignment software package first described by David J. Lipman and William R. Pearson in 1985. Its legacy is the FASTA format which is now ubiquitous in bioinformatics. History The original FASTA program was designed for protein sequence similarity searching. Because of the exponentially expanding genetic information and the limited speed and memory of computers in the 1980s heuristic methods were introduced aligning a query sequence to entire data-bases. FASTA, published in 1987, added the ability to do DNA:DNA searches, translated protein:DNA searches, and also provided a more sophisticated shuffling program for evaluating statistical significance. There are several programs in this package that allow the alignment of protein sequences and DNA sequences. Nowadays, increased computer performance makes it possible to perform searches for local alignment detection in a database using the Smith–Waterman algorithm. FASTA is pronounced "f ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Progressive Strategy
Progressive may refer to: Politics * Progressivism, a political philosophy in support of social reform ** Progressivism in the United States, the political philosophy in the American context * Progressive realism, an American foreign policy paradigm focused on producing measurable results in pursuit of widely supported goals Political organizations * Congressional Progressive Caucus, members within the Democratic Party in the United States Congress dedicated to the advancement of progressive issues and positions * Progressive Alliance (other) * Progressive Conservative (other) * Progressive Party (other) * Progressive Unionist (other) Other uses in politics * Progressive Era, a period of reform in the United States (c. 1890–1930) * Progressive tax, a type of tax rate structure Arts, entertainment, and media Music * Progressive music, a type of music that expands stylistic boundaries outwards * "Progressive" (song), a 2009 single ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MUSCLE (alignment Software)
MUltiple Sequence Comparison by Log-Expectation (MUSCLE) is computer software for multiple sequence alignment of protein and nucleotide sequences. It is licensed as public domain. The method was published by Robert C. Edgar in two papers in 2004. The first paper, published in ''Nucleic Acids Research'', introduced the sequence alignment algorithm. The second paper, published in ''BMC Bioinformatics'', presented more technical details. Algorithm The MUSCLE algorithm proceeds in three stages: the ''draft progressive'', ''improved progressive'', and ''refinement'' stage. Stage 1: Draft Progressive In this first stage, the algorithm produces a multiple alignment, emphasizing speed over accuracy. This step begins by computing the k-mer distance for every pair of input sequences to create a distance matrix. UPGMA clusters the distance matrix to produce a binary tree. From this tree a progressive alignment is constructed, beginning with the creation of profiles for each leaf of the tree ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
MAFFT
In bioinformatics, MAFFT (for multiple alignment using fast Fourier transform) is a program used to create multiple sequence alignments of amino acid or nucleotide sequences. Published in 2002, the first version of MAFFT used an algorithm based on progressive alignment, in which the sequences were clustered with the help of the Fast Fourier Transform. Subsequent versions of MAFFT have added other algorithms and modes of operation, including options for faster alignment of large numbers of sequences, higher accuracy alignments, alignment of non-coding RNA sequences, and the addition of new sequences to existing alignments. See also * Sequence alignment software * Clustal Clustal is a series of widely used computer programs used in bioinformatics for multiple sequence alignment. There have been many versions of Clustal over the development of the algorithm that are listed below. The analysis of each tool and its ... References External links * MAFFT Online ServerMAFFT ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
UNIX
Unix (; trademarked as UNIX) is a family of multitasking, multiuser computer operating systems that derive from the original AT&T Unix, whose development started in 1969 at the Bell Labs research center by Ken Thompson, Dennis Ritchie, and others. Initially intended for use inside the Bell System, AT&T licensed Unix to outside parties in the late 1970s, leading to a variety of both academic and commercial Unix variants from vendors including University of California, Berkeley ( BSD), Microsoft ( Xenix), Sun Microsystems (SunOS/ Solaris), HP/ HPE (HP-UX), and IBM ( AIX). In the early 1990s, AT&T sold its rights in Unix to Novell, which then sold the UNIX trademark to The Open Group, an industry consortium founded in 1996. The Open Group allows the use of the mark for certified operating systems that comply with the Single UNIX Specification (SUS). Unix systems are characterized by a modular design that is sometimes called the " Unix philosophy". According to this p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Information Resource
The Protein Information Resource (PIR), located at Georgetown University Medical Center, is an integrated public bioinformatics resource to support genomic and proteomic research, and scientific studies. It contains protein sequences databases History PIR was established in 1984 by the National Biomedical Research Foundation as a resource to assist researchers and customers in the identification and interpretation of protein sequence information. Prior to that, the foundation compiled the first comprehensive collection of macromolecular sequences in the Atlas of Protein Sequence and Structure, published from 1964 to 1974 under the editorship of Margaret Dayhoff. Dayhoff and her research group pioneered in the development of computer methods for the comparison of protein sequences, for the detection of distantly related sequences and duplications within sequences, and for the inference of evolutionary histories from alignments of protein sequences. Winona Barker and Robert Ledl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Linux
Linux ( or ) is a family of open-source Unix-like operating systems based on the Linux kernel, an operating system kernel first released on September 17, 1991, by Linus Torvalds. Linux is typically packaged as a Linux distribution, which includes the kernel and supporting system software and libraries, many of which are provided by the GNU Project. Many Linux distributions use the word "Linux" in their name, but the Free Software Foundation uses the name "GNU/Linux" to emphasize the importance of GNU software, causing some controversy. Popular Linux distributions include Debian, Fedora Linux, and Ubuntu, the latter of which itself consists of many different distributions and modifications, including Lubuntu and Xubuntu. Commercial distributions include Red Hat Enterprise Linux and SUSE Linux Enterprise. Desktop Linux distributions include a windowing system such as X11 or Wayland, and a desktop environment such as GNOME or KDE Plasma. Distributions intended for ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |




.jpg)