|
Solenoid Protein Domain
Solenoid protein domains are a highly modular type of protein domain. They consist of a chain of nearly identical folds, often simply called tandem repeats. They are extremely common among all types of proteins, though exact figures are unknown. "Repeats" in molecular biology In proteins, a "repeat" is any sequence block that returns more than one time in the sequence, either in an identical or a highly similar form. Repetitiveness does not in itself indicate anything about the structure of the protein. As a "rule of thumb", short repetitive sequences (e.g. those below the length of 10 amino acids) may be intrinsically disordered, and not part of any folded protein domains. Repeats that are at least 30 to 40 amino acids long, are far more likely to be folded as part of a domain. Such long repeats are frequently indicative of the presence of a solenoid domain in the protein. Examples of disordered repetitive sequences include the 7-mer peptide repeats found in the RPB1 subun ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
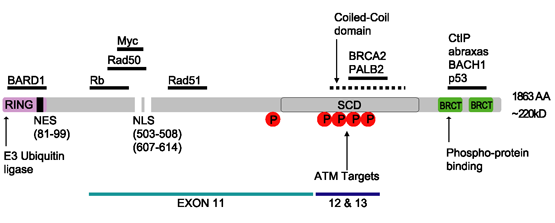
BRCA1
Breast cancer type 1 susceptibility protein is a protein that in humans is encoded by the ''BRCA1'' () gene. Orthologs are common in other vertebrate species, whereas invertebrate genomes may encode a more distantly related gene. ''BRCA1'' is a human tumor suppressor gene (also known as a caretaker gene) and is responsible for repairing DNA. ''BRCA1'' and '' BRCA2'' are unrelated proteins, but both are normally expressed in the cells of breast and other tissue, where they help repair damaged DNA, or destroy cells if DNA cannot be repaired. They are involved in the repair of chromosomal damage with an important role in the error-free repair of DNA double-strand breaks. If ''BRCA1'' or ''BRCA2'' itself is damaged by a BRCA mutation, damaged DNA is not repaired properly, and this increases the risk for breast cancer. ''BRCA1'' and ''BRCA2'' have been described as "breast cancer susceptibility genes" and "breast cancer susceptibility proteins". The predominant allele has a normal, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta Barrel
In protein structures, a beta barrel is a beta sheet composed of tandem repeats that twists and coils to form a closed toroidal structure in which the first strand is bonded to the last strand (hydrogen bond). Beta-strands in many beta-barrels are arranged in an antiparallel fashion. Beta barrel structures are named for resemblance to the barrels used to contain liquids. Most of them are water-soluble proteins and frequently bind hydrophobic ligands in the barrel center, as in lipocalins. Others span cell membranes and are commonly found in porins. Porin-like barrel structures are encoded by as many as 2–3% of the genes in Gram-negative bacteria. It has been shown that more than 600 proteins with various function (e.g., oxidase, dismutase, amylase) contain the beta barrel structure. In many cases, the strands contain alternating polar and non-polar (hydrophilic and hydrophobic) amino acids, so that the hydrophobic residues are oriented into the interior of the barrel to form a hy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Kelch Protein
Kelch proteins (and Kelch-like proteins) are a widespread group of proteins that contain multiple Kelch motifs. The kelch domain generally occurs as a set of five to seven kelch tandem repeats that form a β-propeller tertiary structure. Kelch-repeat β-propellers are generally involved in protein–protein interactions, though the large diversity of domain architectures and limited sequence identity between kelch motifs make characterisation of the kelch superfamily difficult. Structure The N-terminus of several Kelch proteins contain other protein domains, including Discoidin, F-box, and Broad-complex, Tramtrack, Bric-a-Brac/Poxvirus Zinc finger (BTB/POZ) domains. Kelch proteins may also only have a β-propeller architecture. The BTB domain of kelch proteins (if present) allows the formation of homo- or heterodimers that mediate protein–protein interactions. The C-terminus of Kelch proteins contains kelch repeats. Each kelch repeat is a sequence of 44–55 amino acids in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
WD40 Repeat
The WD40 repeat (also known as the WD or beta-transducin repeat) is a short structural motif of approximately 40 amino acids, often terminating in a tryptophan-aspartic acid (W-D) dipeptide. Tandem copies of these repeats typically fold together to form a type of circular solenoid protein domain called the WD40 domain. Structure WD40 domain-containing proteins have 4 to 16 repeating units, all of which are thought to form a circularised beta-propeller structure (see figure to the right). The WD40 domain is composed of several repeats, a variable region of around 20 residues at the beginning followed by a more common repeated set of residues. These repeats typically form a four stranded anti-parallel beta sheet or blade. These blades come together to form a propeller with the most common being a 7 bladed beta propeller. The blades interlock so that the last beta strand of one repeat forms with the first three of the next repeat to form the 3D blade structure. Function WD40-repe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tetratricopeptide
The tetratricopeptide repeat (TPR) is a structural motif. It consists of a Degeneracy (biology), degenerate 34 amino acid protein tandem repeats, tandem repeat identified in a wide variety of proteins. It is found in tandem arrays of 3–16 motifs, which form scaffolds to mediate protein–protein interactions and often the assembly of multiprotein complexes. These alpha-helix pair repeats usually protein folding, fold together to produce a single, linear solenoid protein domain, solenoid domain called a TPR domain. Proteins with such domains include the anaphase-promoting complex (APC) subunits CDC16, cdc16, CDC23, cdc23 and CDC27, cdc27, the NADPH oxidase subunit neutrophil cytosolic factor 2, p67-phox, hsp90-binding immunophilins, transcription factors, the protein kinase R (PKR), the major receptor for peroxisomal matrix protein import PEX5, protein arginine methyltransferase 9 (PRMT9), and mitochondrial import proteins. Structure The structure of the PPP5C, PP5 protein w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Armadillo Repeats
An armadillo repeat is the name of a characteristic, repetitive amino acid sequence of about 40 residues in length that is found in many proteins. Proteins that contain armadillo repeats typically contain several tandemly repeated copies. Each armadillo repeat is composed of a pair of alpha helices that form a hairpin structure. Multiple copies of the repeat form what is known as an alpha solenoid structure. Examples of proteins that contain armadillo repeats include β-catenin, α-importin, plakoglobin, adenomatous polyposis coli (APC), and many others. The term armadillo derives from the historical name of the β-catenin gene in the fruitfly ''Drosophila'' where the armadillo repeat was first discovered. Although β-catenin was previously believed to be a protein involved in linking cadherin cell adhesion proteins to the cytoskeleton, recent work indicates that β-catenin regulates the homodimerization of alpha-catenin, which in turn controls actin branching and bundling.Nuss ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ankyrin Repeat
The ankyrin repeat is a 33-residue motif in proteins consisting of two alpha helices separated by loops, first discovered in signaling proteins in yeast Cdc10 and ''Drosophila'' Notch. Domains consisting of ankyrin tandem repeats mediate protein–protein interactions and are among the most common structural motifs in known proteins. They appear in bacterial, archaeal, and eukaryotic proteins, but are far more common in eukaryotes. Ankyrin repeat proteins, though absent in most viruses, are common among poxviruses. Most proteins that contain the motif have four to six repeats, although its namesake ankyrin contains 24, and the largest known number of repeats is 34, predicted in a protein expressed by ''Giardia lamblia''. Ankyrin repeats typically fold together to form a single, linear solenoid structure called ankyrin repeat domains. These domains are one of the most common protein–protein interaction platforms in nature. They occur in a large number of functionally diverse ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HEAT Repeat Domain
A HEAT repeat is a protein tandem repeat structural motif composed of two alpha helices linked by a short loop. HEAT repeats can form alpha solenoids, a type of solenoid protein domain found in a number of cytoplasmic proteins. The name "HEAT" is an acronym for four proteins in which this repeat structure is found: Huntingtin, elongation factor 3 (EF3), protein phosphatase 2A (PP2A), and the yeast kinase TOR1. HEAT repeats form extended superhelical structures which are often involved in intracellular transport; they are structurally related to armadillo repeats. The nuclear transport protein importin beta contains 19 HEAT repeats. Various HEAT repeat proteins and their structures Representative examples of HEAT repeat proteins include importin β (also known as karyopherin β) family, regulatory subunits of condensin and cohesin, separase, PIKKs (phosphatidylinositol 3-kinase-related protein kinases) such as ATM ( Ataxia telangiectasia mutated) and ATR (Ataxia telangiectasia an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |