|
HEAT Repeat Domain
A HEAT repeat is a protein tandem repeat structural motif composed of two alpha helices linked by a short loop. HEAT repeats can form alpha solenoids, a type of solenoid protein domain found in a number of cytoplasmic proteins. The name "HEAT" is an acronym for four proteins in which this repeat structure is found: Huntingtin, elongation factor 3 (EF3), protein phosphatase 2A (PP2A), and the yeast kinase TOR1. HEAT repeats form extended superhelical structures which are often involved in intracellular transport; they are structurally related to armadillo repeats. The nuclear transport protein importin beta contains 19 HEAT repeats. Various HEAT repeat proteins and their structures Representative examples of HEAT repeat proteins include importin β (also known as karyopherin β) family, regulatory subunits of condensin and cohesin, separase, PIKKs (phosphatidylinositol 3-kinase-related protein kinases) such as ATM ( Ataxia telangiectasia mutated) and ATR (Ataxia telangiectasia an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Phosphatase 2A
Protein phosphatase 2A may refer to: * Protein phosphatase 2 Protein phosphatase 2 (PP2), also known as PP2A, is an enzyme that in humans is encoded by the ''PPP2CA'' gene. The PP2A heterotrimeric protein phosphatase is ubiquitously expressed, accounting for a large fraction of phosphatase activity in eu ..., an enzyme * (myosin-light-chain) phosphatase, an enzyme {{Short pages monitor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cohesin
Cohesin is a protein complex that mediates sister chromatid cohesion, homologous recombination, and DNA looping. Cohesin is formed of SMC3, SMC1, SCC1 and SCC3 ( SA1 or SA2 in humans). Cohesin holds sister chromatids together after DNA replication until anaphase when removal of cohesin leads to separation of sister chromatids. The complex forms a ring-like structure and it is believed that sister chromatids are held together by entrapment inside the cohesin ring. Cohesin is a member of the SMC family of protein complexes which includes Condensin, MukBEF and SMC-ScpAB. Cohesin was separately discovered in budding yeast by Douglas Koshland and Kim Nasmyth. Structure Cohesin is a multi-subunit protein complex, made up of SMC1, SMC3, RAD21 and SCC3 (SA1 or SA2). SMC1 and SMC3 are members of the Structural Maintenance of Chromosomes (SMC) family. SMC proteins have two main structural characteristics: an ATP-binding cassette-like 'head' domain with ATPase activity (form ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Condensin
Condensins are large protein complexes that play a central role in chromosome assembly and segregation during mitosis and meiosis (Figure 1). Their subunits were originally identified as major components of mitotic chromosomes assembled in ''Xenopus'' egg extracts. Subunit composition Eukaryotic types Many eukaryotic cells possess two different types of condensin complexes, known as condensin I and condensin II, each of which is composed of five subunits (Figure 2). Condensins I and II share the same pair of core subunits, SMC2 and SMC4, both belonging to a large family of chromosomal ATPases, known as SMC proteins (SMC stands for Structural Maintenance of Chromosomes). Each of the complexes contains a distinct set of non-SMC regulatory subunits (a kleisin subunit and a pair of HEAT repeat subunits). Both complexes are large, having a total molecular mass of 650-700 kDa. The core subunits condensins (SMC2 and SMC4) are conserved among all eukaryotic species that have been ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cohesin
Cohesin is a protein complex that mediates sister chromatid cohesion, homologous recombination, and DNA looping. Cohesin is formed of SMC3, SMC1, SCC1 and SCC3 ( SA1 or SA2 in humans). Cohesin holds sister chromatids together after DNA replication until anaphase when removal of cohesin leads to separation of sister chromatids. The complex forms a ring-like structure and it is believed that sister chromatids are held together by entrapment inside the cohesin ring. Cohesin is a member of the SMC family of protein complexes which includes Condensin, MukBEF and SMC-ScpAB. Cohesin was separately discovered in budding yeast by Douglas Koshland and Kim Nasmyth. Structure Cohesin is a multi-subunit protein complex, made up of SMC1, SMC3, RAD21 and SCC3 (SA1 or SA2). SMC1 and SMC3 are members of the Structural Maintenance of Chromosomes (SMC) family. SMC proteins have two main structural characteristics: an ATP-binding cassette-like 'head' domain with ATPase activity (form ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FANCF
Fanconi anemia group F protein is a protein that in humans is encoded by the ''FANCF'' gene. Interactions FANCF has been shown to interact with Fanconi anemia, complementation group C, FANCG, FANCA and FANCE. Function FANCF is an adaptor protein that plays a key role in the proper assembly of the FA core complex. The FA core complex is composed of eight proteins (FANCA, -B, -C, -E, -F, -G, -L and -M). FANCF stabilizes the interaction between the FANCC/FANCE subcomplex and the FANCA/FANCG subcomplex and locks the whole FA core complex in a conformation that is essential to perform its function in DNA repair. The FA core complex is a nuclear core complex that is essential for the monoubiquitination of FANCD2 and this modified form of FANCD2 colocalizes with BRCA1, RAD51 and PCNA in foci that also contain other DNA repair proteins. All these proteins function together to facilitate DNA interstrand cross-link repair. They also function in other DNA damage response repair proce ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Fanconi Anemia
Fanconi anaemia (FA) is a rare genetic disease resulting in impaired response to DNA damage. Although it is a very rare disorder, study of this and other bone marrow failure syndromes has improved scientific understanding of the mechanisms of normal bone marrow function and development of cancer. Among those affected, the majority develop cancer, most often acute myelogenous leukemia (AML), and 90% develop aplastic anemia (the inability to produce blood cells) by age 40. About 60–75% have congenital defects, commonly short stature, abnormalities of the skin, arms, head, eyes, kidneys, and ears, and developmental disabilities. Around 75% have some form of endocrine problem, with varying degrees of severity. FA is the result of a genetic defect in a cluster of proteins responsible for DNA repair via homologous recombination. Treatment with androgens and hematopoietic (blood cell) growth factors can help bone marrow failure temporarily, but the long-term treatment is bone marrow ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA-PKcs
DNA-dependent protein kinase, catalytic subunit, also known as DNA-PKcs, is an enzyme that in humans is encoded by the gene designated as ''PRKDC'' or ''XRCC7''. DNA-PKcs belongs to the phosphatidylinositol 3-kinase-related kinase protein family. The DNA-Pkcs protein is a serine/threonine protein kinase comprising a single polypeptide chain of 4,128 amino acids. Function DNA-PKcs is the catalytic subunit of a nuclear DNA-dependent serine/threonine protein kinase called DNA-PK. The second component is the autoimmune antigen Ku. On its own, DNA-PKcs is inactive and relies on Ku to direct it to DNA ends and trigger its kinase activity. DNA-PKcs is required for the non-homologous end joining (NHEJ) pathway of DNA repair, which rejoins double-strand breaks. It is also required for V(D)J recombination, a process that utilizes NHEJ to promote immune system diversity. DNA-PKcs knockout mice have severe combined immunodeficiency due to their V(D)J recombination defect. Many proteins ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
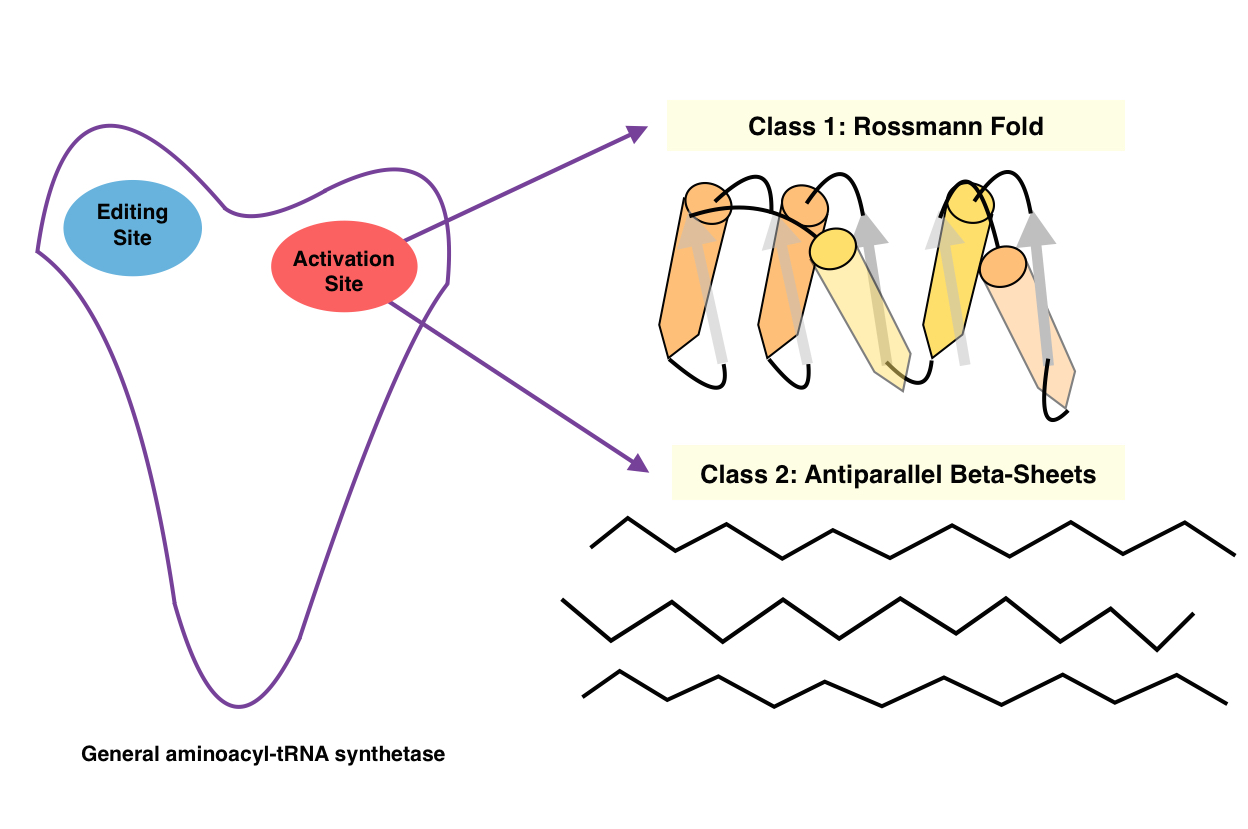
Aminoacyl TRNA Synthetase
An aminoacyl-tRNA synthetase (aaRS or ARS), also called tRNA-ligase, is an enzyme that attaches the appropriate amino acid onto its corresponding tRNA. It does so by catalyzing the transesterification of a specific cognate amino acid or its precursor to one of all its compatible cognate tRNAs to form an aminoacyl-tRNA. In humans, the 20 different types of aa-tRNA are made by the 20 different aminoacyl-tRNA synthetases, one for each amino acid of the genetic code. This is sometimes called "charging" or "loading" the tRNA with an amino acid. Once the tRNA is charged, a ribosome can transfer the amino acid from the tRNA onto a growing peptide, according to the genetic code. Aminoacyl tRNA therefore plays an important role in RNA translation, the expression of genes to create proteins. Mechanism The synthetase first binds ATP and the corresponding amino acid (or its precursor) to form an aminoacyl-adenylate, releasing inorganic pyrophosphate (PPi). The adenylate-aaRS complex then ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Translational Regulation
Translational regulation refers to the Regulation of gene expression, control of the levels of protein synthesized from its mRNA. This regulation is vastly important to the cellular response to stressors, growth cues, and Cellular differentiation, differentiation. In comparison to transcriptional regulation, it results in much more immediate cellular adjustment through direct regulation of protein concentration. The corresponding mechanisms are primarily targeted on the control of ribosome recruitment on the initiation codon, but can also involve modulation of peptide elongation, termination of protein synthesis, or ribosome biogenesis. While these general concepts are widely conserved, some of the finer details in this sort of regulation have been proven to differ between prokaryotic and eukaryotic organisms. In prokaryotes Initiation Initiation of translation is regulated by the accessibility of ribosomes to the Shine-Dalgarno sequence. This stretch of four to nine purine resi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Transcriptional Regulation
In molecular biology and genetics, transcriptional regulation is the means by which a cell regulates the conversion of DNA to RNA (transcription), thereby orchestrating gene activity. A single gene can be regulated in a range of ways, from altering the number of copies of RNA that are transcribed, to the temporal control of when the gene is transcribed. This control allows the cell or organism to respond to a variety of intra- and extracellular signals and thus mount a response. Some examples of this include producing the mRNA that encode enzymes to adapt to a change in a food source, producing the gene products involved in cell cycle specific activities, and producing the gene products responsible for cellular differentiation in multicellular eukaryotes, as studied in evolutionary developmental biology. The regulation of transcription is a vital process in all living organisms. It is orchestrated by transcription factors and other proteins working in concert to finely tune t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
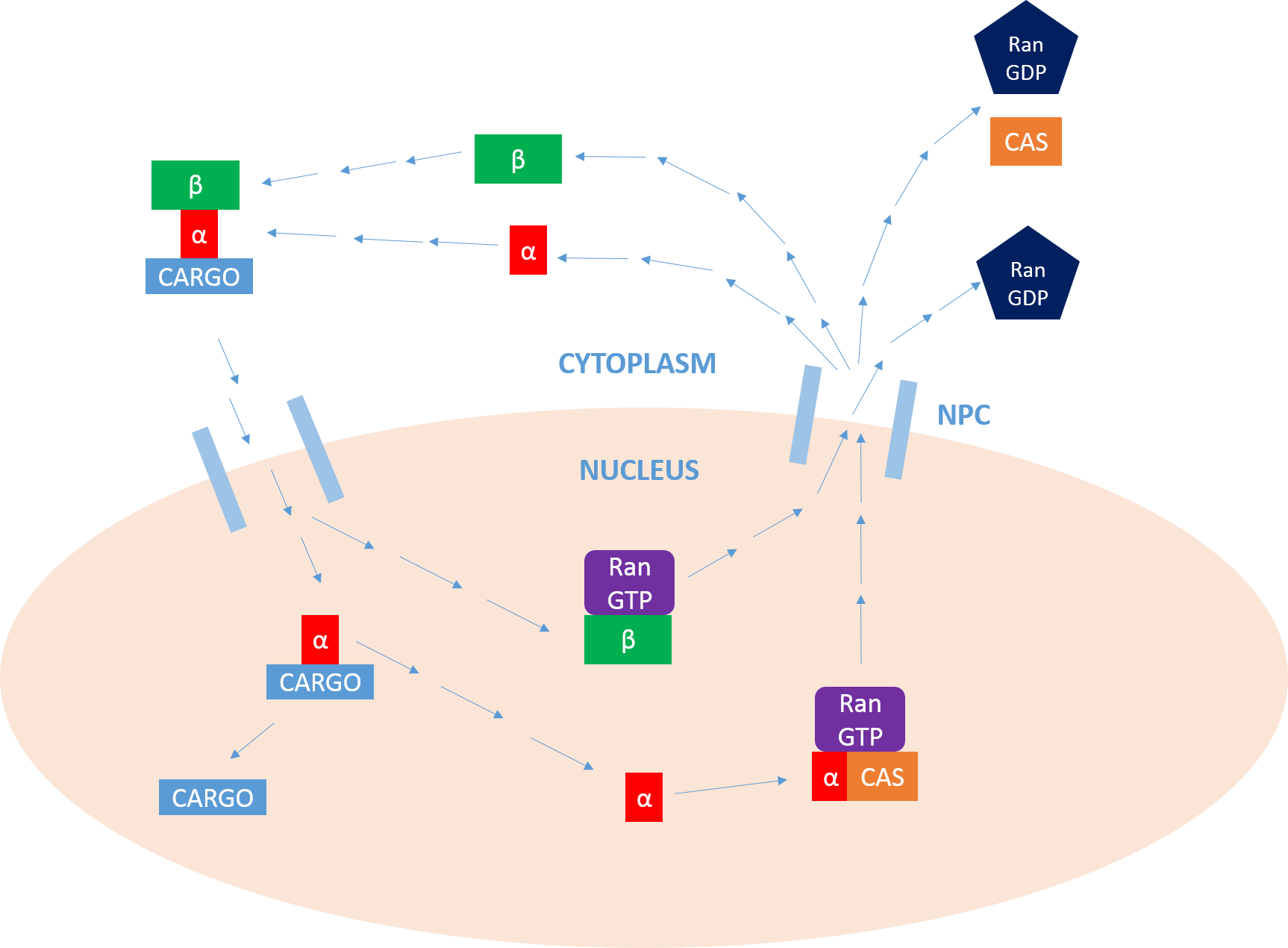
Importin
Importin is a type of karyopherin that transports protein molecules from the cell's cytoplasm to the nucleus. It does so by binding to specific recognition sequences, called nuclear localization sequences (NLS). Importin has two subunits, importin α and importin β. Members of the importin-β family can bind and transport cargo by themselves, or can form heterodimers with importin-α. As part of a heterodimer, importin-β mediates interactions with the pore complex, while importin-α acts as an adaptor protein to bind the nuclear localization signal (NLS) on the cargo. The NLS-Importin α-Importin β trimer dissociates after binding to Ran GTP inside the nucleus, with the two importin proteins being recycled to the cytoplasm for further use. Discovery Importin can exist as either a heterodimer of importin-α/β or as a monomer of Importin-β. Importin-α was first isolated in 1994 by a group includinEnno Hartmann based at the Max Delbrück Center for Molecular Medicine. The p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Proteasome
Proteasomes are protein complexes which degrade unneeded or damaged proteins by proteolysis, a chemical reaction that breaks peptide bonds. Enzymes that help such reactions are called proteases. Proteasomes are part of a major mechanism by which cells regulate the concentration of particular proteins and degrade misfolded proteins. Proteins are tagged for degradation with a small protein called ubiquitin. The tagging reaction is catalyzed by enzymes called ubiquitin ligases. Once a protein is tagged with a single ubiquitin molecule, this is a signal to other ligases to attach additional ubiquitin molecules. The result is a ''polyubiquitin chain'' that is bound by the proteasome, allowing it to degrade the tagged protein. The degradation process yields peptides of about seven to eight amino acids long, which can then be further degraded into shorter amino acid sequences and used in synthesizing new proteins. Proteasomes are found inside all eukaryotes and archaea, and in so ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |